Google DeepMind Releases AlphaGenome: A Deep Learning Model that can more Comprehensively Predict the Impact of Single Variants or Mutations in DNA
A Unified Deep Learning Model to Understand the Genome Google DeepMind has unveiled AlphaGenome, a new deep learning framework designed to predict the regulatory consequences of DNA sequence variations across a wide spectrum of biological modalities. AlphaGenome stands out by accepting long DNA sequences—up to 1 megabase—and outputting high-resolution predictions, such as base-level splicing events, […] The post Google DeepMind Releases AlphaGenome: A Deep Learning Model that can more Comprehensively Predict the Impact of Single Variants or Mutations in DNA appeared first on MarkTechPost.

A Unified Deep Learning Model to Understand the Genome
Google DeepMind has unveiled AlphaGenome, a new deep learning framework designed to predict the regulatory consequences of DNA sequence variations across a wide spectrum of biological modalities. AlphaGenome stands out by accepting long DNA sequences—up to 1 megabase—and outputting high-resolution predictions, such as base-level splicing events, chromatin accessibility, gene expression, and transcription factor binding.
Built to address limitations in earlier models, AlphaGenome bridges the gap between long-sequence input processing and nucleotide-level output precision. It unifies predictive tasks across 11 output modalities and handles over 5,000 human genomic tracks and 1,000+ mouse tracks. This level of multimodal capability positions AlphaGenome as one of the most comprehensive sequence-to-function models in genomics.
Technical Architecture and Training Methodology
AlphaGenome adopts a U-Net-style architecture with a transformer core. It processes DNA sequences in 131kb parallelized chunks across TPUv3 devices, enabling context-aware, base-pair-resolution predictions. The architecture uses two-dimensional embeddings for spatial interaction modeling (e.g., contact maps) and one-dimensional embeddings for linear genomic tasks.
Training involved two stages:
- Pre-training: using fold-specific and all-folds models to predict from observed experimental tracks.
- Distillation: a student model learns from teacher models to deliver consistent and efficient predictions, enabling fast inference (~1 second per variant) on GPUs like the NVIDIA H100.

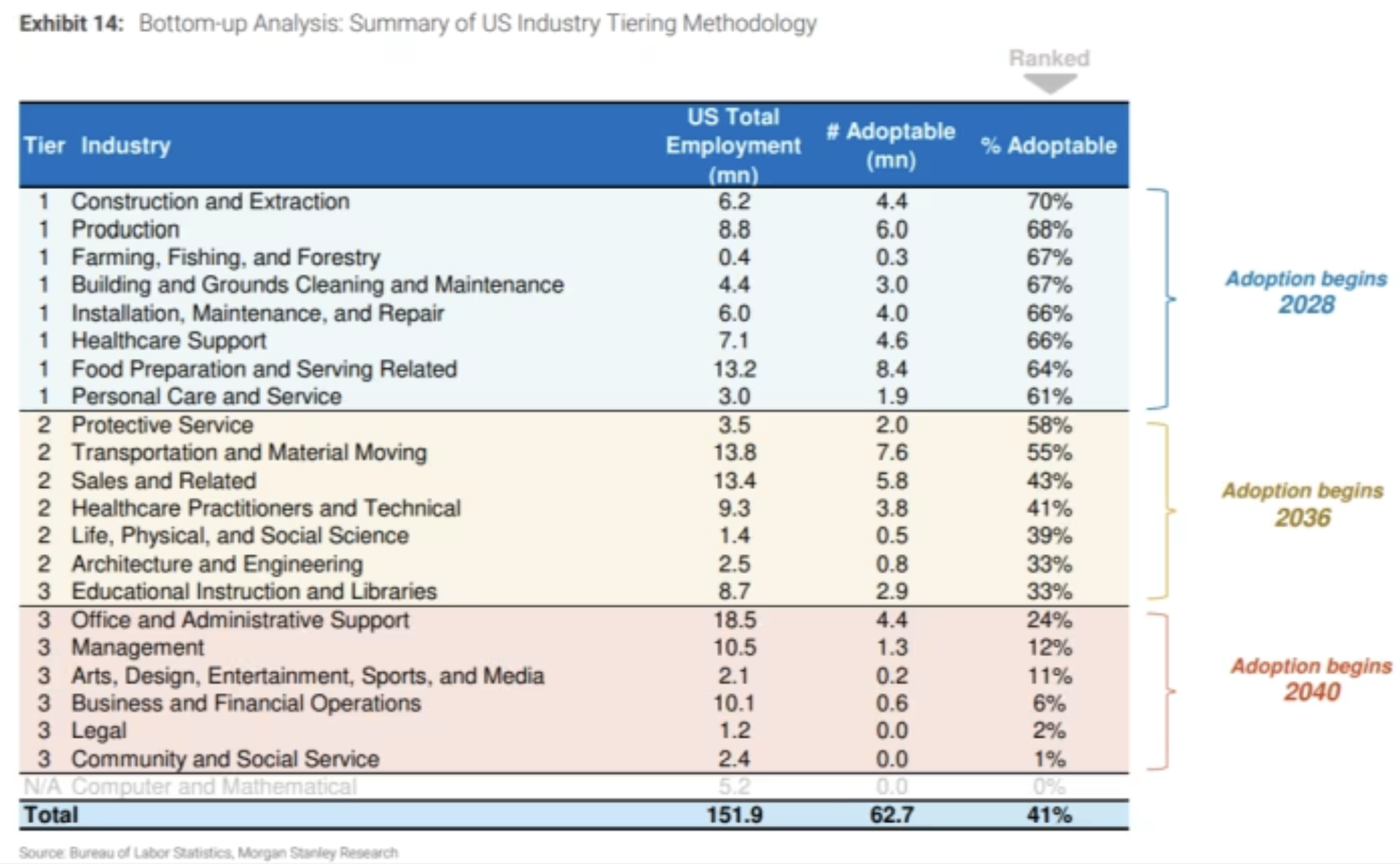
Performance Across Benchmarks
AlphaGenome was rigorously benchmarked against specialized and multimodal models across 24 genome track and 26 variant effect prediction tasks. It outperformed or matched state-of-the-art models in 22/24 and 24/26 evaluations, respectively. In splicing, gene expression, and chromatin-related tasks, it consistently surpassed specialized models like SpliceAI, Borzoi, and ChromBPNet.
For instance:
- Splicing: AlphaGenome is the first to simultaneously model splice sites, splice site usage, and splice junctions at 1 bp resolution. It outperformed Pangolin and SpliceAI on 6 of 7 benchmarks.
- eQTL prediction: The model achieved a 25.5% relative improvement in direction-of-effect prediction compared to Borzoi.
- Chromatin accessibility: It demonstrated strong correlation with DNase-seq and ATAC-seq experimental data, outperforming ChromBPNet by 8-19%.

Variant Effect Prediction from Sequence Alone
One of AlphaGenome’s key strengths lies in variant effect prediction (VEP). It handles zero-shot and supervised VEP tasks without relying on population genetics data, making it robust for rare variants and distal regulatory regions. With a single inference, AlphaGenome evaluates how a mutation may impact splicing patterns, expression levels, and chromatin state—all in a multimodal fashion.
The model’s ability to reproduce clinically observed splicing disruptions, such as exon skipping or novel junction formation, illustrates its utility in diagnosing rare genetic diseases. It accurately modeled the effects of a 4bp deletion in the DLG1 gene observed in GTEx samples.
Application in GWAS Interpretation and Disease Variant Analysis
AlphaGenome aids in interpreting GWAS signals by assigning directionality of variant effects on gene expression. Compared to colocalization methods like COLOC, AlphaGenome provided complementary and broader coverage—resolving 4x more loci in the lowest MAF quintile.
It also demonstrated utility in cancer genomics. When analyzing non-coding mutations upstream of the TAL1 oncogene (linked to T-ALL), AlphaGenome’s predictions matched known epigenomic changes and expression upregulation mechanisms, confirming its ability to assess gain-of-function mutations in regulatory elements.
TL;DR
AlphaGenome by Google DeepMind is a powerful deep learning model that predicts the effects of DNA mutations across multiple regulatory modalities at base-pair resolution. It combines long-range sequence modeling, multimodal prediction, and high-resolution output in a unified architecture. Outperforming specialized and generalist models across 50 benchmarks, AlphaGenome significantly improves the interpretation of non-coding genetic variants and is now available in preview to support genomics research worldwide.
Check out the Paper, Technical details and GitHub Page. All credit for this research goes to the researchers of this project. Also, feel free to follow us on Twitter and don’t forget to join our 100k+ ML SubReddit and Subscribe to our Newsletter.
The post Google DeepMind Releases AlphaGenome: A Deep Learning Model that can more Comprehensively Predict the Impact of Single Variants or Mutations in DNA appeared first on MarkTechPost.



























































































































































![[The AI Show Episode 156]: AI Answers - Data Privacy, AI Roadmaps, Regulated Industries, Selling AI to the C-Suite & Change Management](https://www.marketingaiinstitute.com/hubfs/ep%20156%20cover.png)
![[The AI Show Episode 155]: The New Jobs AI Will Create, Amazon CEO: AI Will Cut Jobs, Your Brain on ChatGPT, Possible OpenAI-Microsoft Breakup & Veo 3 IP Issues](https://www.marketingaiinstitute.com/hubfs/ep%20155%20cover.png)




































































































![[DEALS] 1min.AI: Lifetime Subscription (82% off) & Other Deals Up To 98% Off – Offers End Soon!](https://www.javacodegeeks.com/wp-content/uploads/2012/12/jcg-logo.jpg)



































































































































_incamerastock_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)
_Brain_light_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)







































































































![Mercedes, Audi, Volvo Reject Apple's New CarPlay Ultra [Report]](https://www.iclarified.com/images/news/97711/97711/97711-640.jpg)