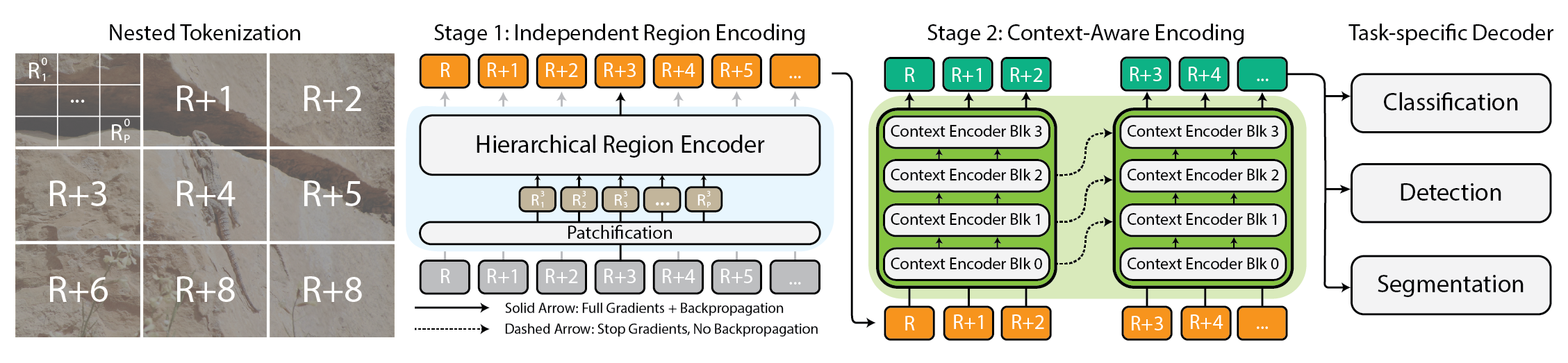
# ProtBFN: Bayesian Foundation Model for Protein Sequence Design
A recent paper published in Nature Communications introduces ProtBFN, a powerful 650 million-parameter foundation model for protein sequence design. ProtBFN uses Bayesian Flow Networks to generate diverse, structurally coherent sequences without relying on explicit structural data oai_citation:0‡nature.com. What ProtBFN Offers Unconditional and conditional protein generation Outperforms leading autoregressive and diffusion models Produces sequences matching natural length and amino acid distributions Includes AbBFN, a fine-tuned variant for antibody heavy chains demonstrated on Observed Antibody Space (OAS) Example Usage from protbfn import ProtBFN import torch model = ProtBFN.load_pretrained("protbfn_650m") seq = model.generate_unconditional(length=150) print(seq) ab_model = ProtBFN.load_pretrained("abbfn_heavy") ab_seq = ab_model.generate_conditional(context="EVQLVESGGGLVQPGG") print(ab_seq) Why It Matters Designing functional proteins often requires intricate modeling of sequence landscapes. ProtBFN’s foundation model approach enables zero-shot design—producing valid proteins without retraining. Its ability to handle both general proteins and antibody-specific sequences makes it versatile for therapeutic and industrial enzyme design. By using probabilistic flow networks, ProtBFN provides both generative flexibility and structural consistency, aligning with core needs of protein engineering. What’s Next The model is open-source and pip-installable. Investigators can benchmark ProtBFN on custom tasks like stability prediction, binding design, or de novo therapeutic protein creation. Community contributions to expand pretrained variants and evaluation metrics are already underway. Sources https://www.nature.com/articles/s41467-025-58250-2 https://github.com/protbfn/protbfn

A recent paper published in Nature Communications introduces ProtBFN, a powerful 650 million-parameter foundation model for protein sequence design. ProtBFN uses Bayesian Flow Networks to generate diverse, structurally coherent sequences without relying on explicit structural data oai_citation:0‡nature.com.
What ProtBFN Offers
- Unconditional and conditional protein generation
- Outperforms leading autoregressive and diffusion models
- Produces sequences matching natural length and amino acid distributions
- Includes AbBFN, a fine-tuned variant for antibody heavy chains demonstrated on Observed Antibody Space (OAS)
Example Usage
from protbfn import ProtBFN
import torch
model = ProtBFN.load_pretrained("protbfn_650m")
seq = model.generate_unconditional(length=150)
print(seq)
ab_model = ProtBFN.load_pretrained("abbfn_heavy")
ab_seq = ab_model.generate_conditional(context="EVQLVESGGGLVQPGG")
print(ab_seq)
Why It Matters
Designing functional proteins often requires intricate modeling of sequence landscapes. ProtBFN’s foundation model approach enables zero-shot design—producing valid proteins without retraining. Its ability to handle both general proteins and antibody-specific sequences makes it versatile for therapeutic and industrial enzyme design.
By using probabilistic flow networks, ProtBFN provides both generative flexibility and structural consistency, aligning with core needs of protein engineering.
What’s Next
The model is open-source and pip-installable. Investigators can benchmark ProtBFN on custom tasks like stability prediction, binding design, or de novo therapeutic protein creation. Community contributions to expand pretrained variants and evaluation metrics are already underway.
Sources
https://www.nature.com/articles/s41467-025-58250-2
https://github.com/protbfn/protbfn






























































































































































![[The AI Show Episode 156]: AI Answers - Data Privacy, AI Roadmaps, Regulated Industries, Selling AI to the C-Suite & Change Management](https://www.marketingaiinstitute.com/hubfs/ep%20156%20cover.png)
![[The AI Show Episode 155]: The New Jobs AI Will Create, Amazon CEO: AI Will Cut Jobs, Your Brain on ChatGPT, Possible OpenAI-Microsoft Breakup & Veo 3 IP Issues](https://www.marketingaiinstitute.com/hubfs/ep%20155%20cover.png)






































































































![Rust VS Go VS TypeScript – which back end language is for you? With Tai Groot [Podcast #176]](https://cdn.hashnode.com/res/hashnode/image/upload/v1750974265013/73f79068-0087-4c39-8a8b-feea8cac873b.png?#)










































































































.jpg?width=1920&height=1920&fit=bounds&quality=70&format=jpg&auto=webp#)