# SandboxAQ Releases Massive Synthetic Molecule Dataset to Accelerate AI Drug Discovery
SandboxAQ, an AI and quantum tech startup spun out of Alphabet and backed by Nvidia, has released a large-scale dataset of synthetic 3D molecular structures aimed at revolutionizing small molecule drug discovery. With over 5.2 million conformers representing diverse chemical scaffolds, the dataset is designed to help researchers train and validate machine learning models capable of predicting drug-target interactions and pharmacological properties. This dataset includes not only optimized 3D geometries but also labels such as protein binding affinity, chemical class, and structural metadata derived from physics-based simulations and available experimental records. The goal is to create a gold standard training resource for geometric deep learning models in drug design. What’s Inside the Dataset Each entry in the dataset includes: 3D atomic coordinates in SDF format SMILES strings and InChIKeys Protein target annotations (when available) Simulated and experimental binding scores Molecular descriptors and class labels SandboxAQ collaborated with pharma partners and used advanced quantum and AI simulations to generate accurate 3D conformers. These molecules were further filtered for drug-likeness, diversity, and relevance to common therapeutic targets in oncology, neurology, and immunology. Developer Integration Developers and researchers can integrate the dataset with graph neural networks, transformer-based models, and molecular docking software. The files are structured for compatibility with frameworks like PyTorch Geometric, DeepChem, and RDKit. Sample Usage from sandboxaq_loader import MoleculeDataset dataset = MoleculeDataset("sandboxaq_5m.sdf") for mol in dataset: print(mol.smiles, mol.binding_score) Modelers can train regression or classification models to predict bioactivity, solubility, or ADMET properties. By using the structural information embedded in 3D coordinates, AI systems can learn features that generalize across diverse chemical families. Why This Matters The release marks one of the largest publicly available synthetic datasets for AI-powered drug discovery. In an industry where early-stage research is slowed by high failure rates and expensive experiments, synthetic data is becoming critical. Models trained on SandboxAQ’s dataset can reduce false positives and help prioritize promising leads, even before any physical assays are performed. By combining AI-generated structures with high-quality annotations, SandboxAQ provides a blueprint for how simulation and data-driven modeling can coexist. The result is a faster, cheaper, and more scalable approach to identifying new drug candidates. Next Steps and Community Access The dataset is available now for academic and non-commercial research, with plans to expand access to industry users via subscription. SandboxAQ has also announced upcoming benchmarks for protein-ligand prediction tasks based on this dataset. As the community begins building and testing models using this resource, SandboxAQ invites contributions, feedback, and proposals for collaborative validation studies. Sources https://www.reuters.com/business/healthcare-pharmaceuticals/nvidia-backed-ai-startup-sandboxaq-creates-new-data-speed-up-drug-discovery-2025-06-18/ https://www.sandboxaq.com/press/2025-drug-discovery-dataset-release

SandboxAQ, an AI and quantum tech startup spun out of Alphabet and backed by Nvidia, has released a large-scale dataset of synthetic 3D molecular structures aimed at revolutionizing small molecule drug discovery. With over 5.2 million conformers representing diverse chemical scaffolds, the dataset is designed to help researchers train and validate machine learning models capable of predicting drug-target interactions and pharmacological properties.
This dataset includes not only optimized 3D geometries but also labels such as protein binding affinity, chemical class, and structural metadata derived from physics-based simulations and available experimental records. The goal is to create a gold standard training resource for geometric deep learning models in drug design.
What’s Inside the Dataset
Each entry in the dataset includes:
- 3D atomic coordinates in SDF format
- SMILES strings and InChIKeys
- Protein target annotations (when available)
- Simulated and experimental binding scores
- Molecular descriptors and class labels
SandboxAQ collaborated with pharma partners and used advanced quantum and AI simulations to generate accurate 3D conformers. These molecules were further filtered for drug-likeness, diversity, and relevance to common therapeutic targets in oncology, neurology, and immunology.
Developer Integration
Developers and researchers can integrate the dataset with graph neural networks, transformer-based models, and molecular docking software. The files are structured for compatibility with frameworks like PyTorch Geometric, DeepChem, and RDKit.
Sample Usage
from sandboxaq_loader import MoleculeDataset
dataset = MoleculeDataset("sandboxaq_5m.sdf")
for mol in dataset:
print(mol.smiles, mol.binding_score)
Modelers can train regression or classification models to predict bioactivity, solubility, or ADMET properties. By using the structural information embedded in 3D coordinates, AI systems can learn features that generalize across diverse chemical families.
Why This Matters
The release marks one of the largest publicly available synthetic datasets for AI-powered drug discovery. In an industry where early-stage research is slowed by high failure rates and expensive experiments, synthetic data is becoming critical. Models trained on SandboxAQ’s dataset can reduce false positives and help prioritize promising leads, even before any physical assays are performed.
By combining AI-generated structures with high-quality annotations, SandboxAQ provides a blueprint for how simulation and data-driven modeling can coexist. The result is a faster, cheaper, and more scalable approach to identifying new drug candidates.
Next Steps and Community Access
The dataset is available now for academic and non-commercial research, with plans to expand access to industry users via subscription. SandboxAQ has also announced upcoming benchmarks for protein-ligand prediction tasks based on this dataset.
As the community begins building and testing models using this resource, SandboxAQ invites contributions, feedback, and proposals for collaborative validation studies.
Sources
https://www.reuters.com/business/healthcare-pharmaceuticals/nvidia-backed-ai-startup-sandboxaq-creates-new-data-speed-up-drug-discovery-2025-06-18/
https://www.sandboxaq.com/press/2025-drug-discovery-dataset-release
































































































































































![[The AI Show Episode 154]: AI Answers: The Future of AI Agents at Work, Building an AI Roadmap, Choosing the Right Tools, & Responsible AI Use](https://www.marketingaiinstitute.com/hubfs/ep%20154%20cover.png)
![[The AI Show Episode 153]: OpenAI Releases o3-Pro, Disney Sues Midjourney, Altman: “Gentle Singularity” Is Here, AI and Jobs & News Sites Getting Crushed by AI Search](https://www.marketingaiinstitute.com/hubfs/ep%20153%20cover.png)






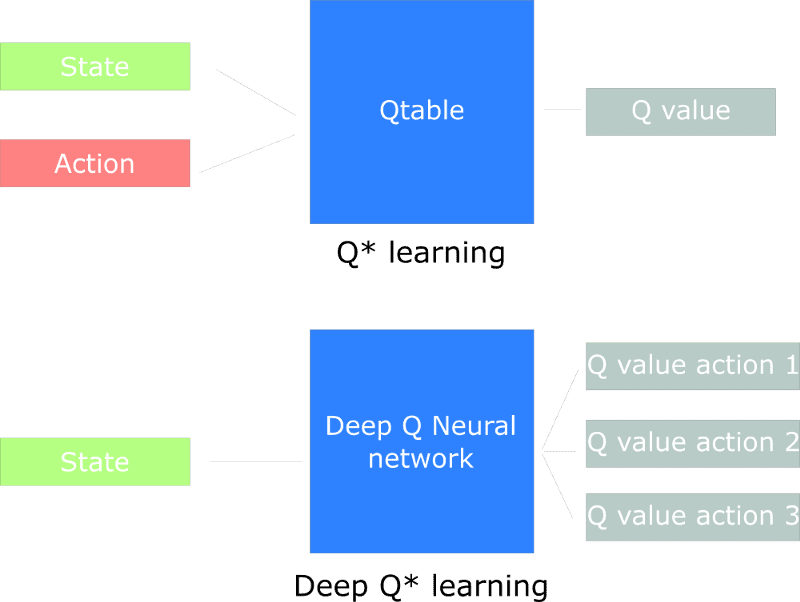
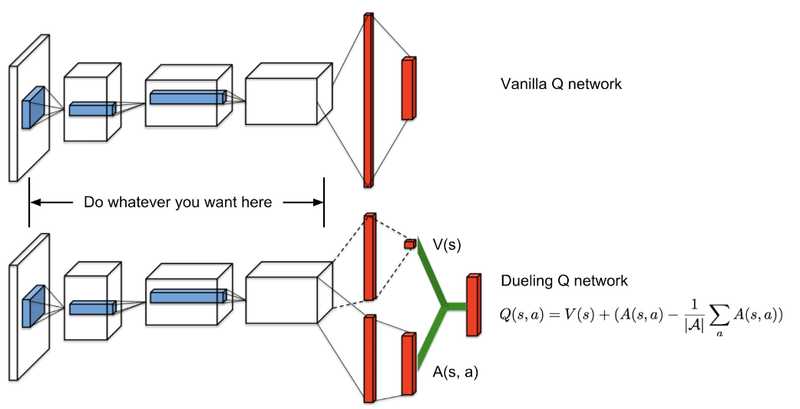
































































































































































![GrandChase tier list of the best characters available [June 2025]](https://media.pocketgamer.com/artwork/na-33057-1637756796/grandchase-ios-android-3rd-anniversary.jpg?#)














































.png?width=1920&height=1920&fit=bounds&quality=70&format=jpg&auto=webp#)





![Apple Weighs Acquisition of AI Startup Perplexity in Internal Talks [Report]](https://www.iclarified.com/images/news/97674/97674/97674-640.jpg)














_Frank_Peters_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)




























































































![Samsung’s Galaxy Z Fold 7 looks positively svelte in dummy hands-on [Video]](https://i0.wp.com/9to5google.com/wp-content/uploads/sites/4/2025/06/Worlds-First-Look-Hands-On-with-the-Galaxy-Z-Fold-7-dummy-1-1-screenshot.png?resize=1200%2C628&quality=82&strip=all&ssl=1)









![Oakley and Meta Launch Smart Glasses for Athletes With AI, 3K Camera, More [Video]](https://www.iclarified.com/images/news/97665/97665/97665-640.jpg)

![How to Get Your Parents to Buy You a Mac, According to Apple [Video]](https://www.iclarified.com/images/news/97671/97671/97671-640.jpg)












![New accessibility settings announced for Steam Big Picture Mode and SteamOS [Beta]](https://www.ghacks.net/wp-content/uploads/2025/06/New-accessibility-settings-announced-for-Steam-Big-Picture-Mode-and-SteamOS.jpg)