New AI Tool Unlocks Hidden Proteins in Sequencing Breakthrough
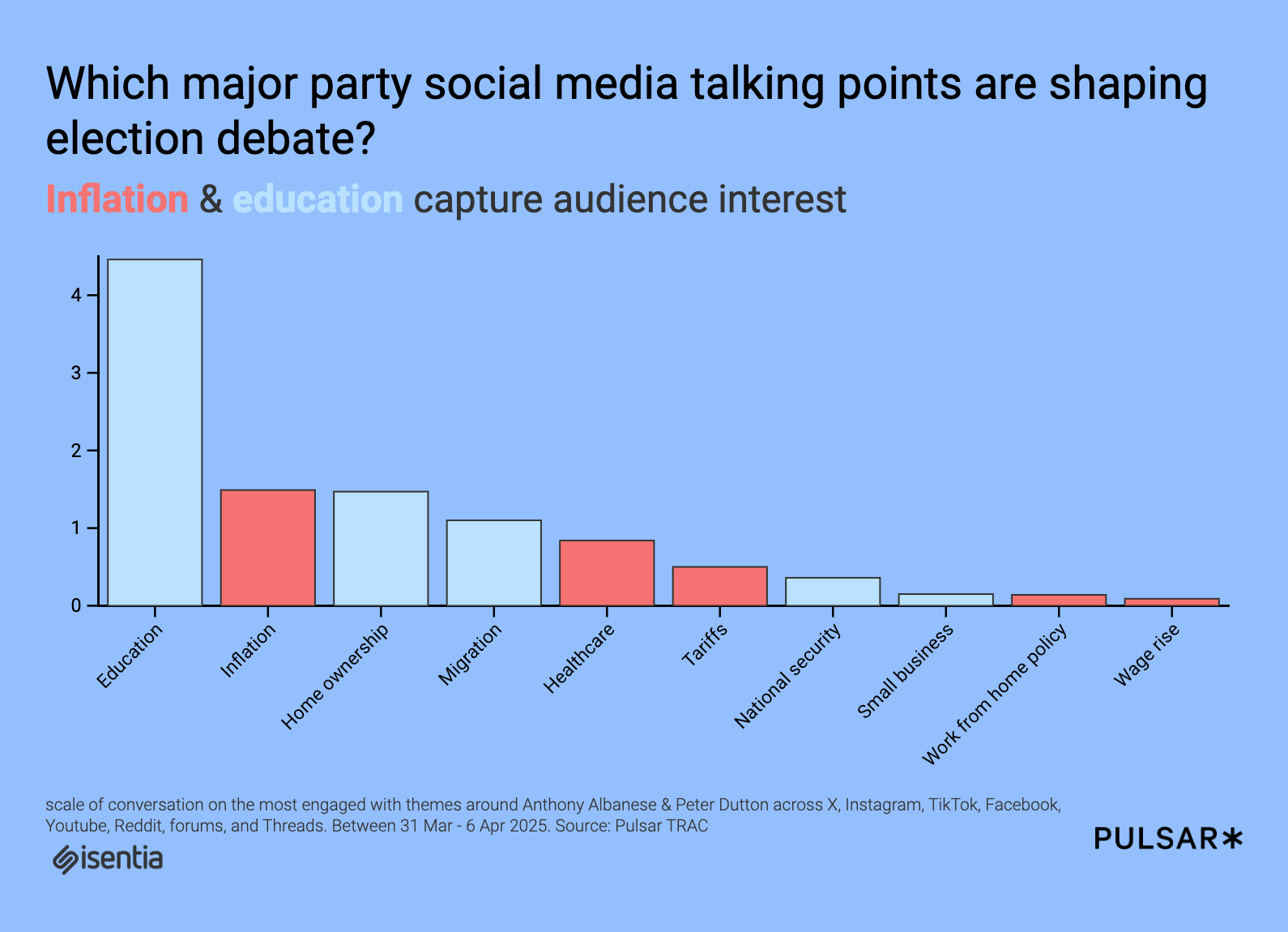
Artificial intelligence (AI) has been hugely instrumental in the field of science. While words like "revolutionary" are often overused, in the case of AI’s role in scientific research, it truly fits. From uncovering new insights in Physics to the discovery of new viruses from dark matter, GenAI is transforming how scientists decode the mysteries of the universe. AI has been particularly groundbreaking in helping scientists understand protein structures - the workhorses of living cells. The technology has now taken a massive step forward with InstaNovo, a new tool designed to advance protein sequencing. InstaNovo holds the potential to uncover more effective cancer treatments, help improve doctors’ understanding of rare diseases, and pave the way for more groundbreaking scientific discoveries in proteomics. Protein sequencing is a long-standing challenge, widely regarded as one of biology's toughest problems. Unlike DNA, which consists of just four bases and follows a relatively simple sequencing method, proteins are composed of 20 amino acids arranged in endless combinations. Even small proteins can display staggering complexity. Adding to the complexity, a protein's function can change entirely depending on how it folds into three-dimensional shapes. Many proteins also undergo chemical modifications after they are formed, which makes it incredibly difficult to trace these changes back to their original genetic blueprint. To address these challenges, InstaNovo was developed as an AI-powered tool designed specifically for de novo protein sequencing. The term de novo refers to rebuilding protein sequences from scratch, rather than relying on existing reference databases. In a paper published in Nature Machine Intelligence, the researchers reveal that InstaNovo is able to leverage AI to reconstruct peptide sequences from scratch, even for proteins that have not been analyzed before. Its breakthrough lies in its ability to decode fragmented peptide signals using a tailored deep-learning method delivering unprecedented efficiency and accuracy. The InstaNovo+ model goes even further. It uses an iterative refinement process that aligns the peptide sequence more closely with spectral data. This is useful in the detection of chemically modified or hidden proteins. The new AI tool is the result of a joint effort between InstaDeep, an AI company, and the Department of Biotechnology and Biomedicine at the Technical University of Denmark (DTU). Key contributors from DTU include Associate Professor Timothy Patrick Jenkins and Assistant Professor Konstantinos Kalogeropoulos. The developers claim that the new tool could revolutionize protein sequencing, much like AlphaFold transformed protein structure prediction. In recognition of its impact, AlphaFold's creators were honored with the Nobel Prize in Chemistry in 2024 for their groundbreaking contributions to the field using AI. “Together, our results and those of others show that scale is the most determining factor in de novo peptide sequencing model performance, as with other fields where the transformer architecture was employed," shared the researchers. “We expect to further increase model performance by taking advantage of the vast amount of MS datasets available in repositories. We also anticipate widespread adoption by peers, and look forward to further exploration of fine-tuning, protein inference, and assembly, as well as building applications on top of our base model for hybrid or de novo searches.” The creation of InstaNovo is not the first attempt by researchers to apply machine learning to protein sequencing. Earlier tools like the AI transformer protein decoder Casanovo showed how AI could help with protein sequencing, but they had a key limitation. Most of them depended heavily on reference databases, which made it hard to identify new or unique proteins. The InstaNovo creators claim that their tool outperforms Casanovo in identifying peptide-spectrum matches (PSMs). The InstaNovo and InstaNovo+ identify 41.8% more PSMs than Casanovo, showcasing their superior capability in complex sequencing tasks. “By eliminating dependency on protein databases and improving accuracy through iterative refinement, InstaNovo and InstaNovo+ uncover previously inaccessible proteomic landscapes, with the potential to drive discoveries across multiple scientific domains,” shared InstaDeep in a blog post. The ability to go “database-free” is not just a side benefit, it is central to what makes InstaNovo innovative. However, the researchers admit that integrating the tool with existing laboratory workflows would be challenging. They also acknowledge that the outputs may require additional verification. Nevertheless, the tools represent a significant step forward in protein research. With further refinement and more real-world testing, the tool can be useful in broadening our understanding of complex biological systems.

Artificial intelligence (AI) has been hugely instrumental in the field of science. While words like "revolutionary" are often overused, in the case of AI’s role in scientific research, it truly fits. From uncovering new insights in Physics to the discovery of new viruses from dark matter, GenAI is transforming how scientists decode the mysteries of the universe.
AI has been particularly groundbreaking in helping scientists understand protein structures - the workhorses of living cells. The technology has now taken a massive step forward with InstaNovo, a new tool designed to advance protein sequencing.
InstaNovo holds the potential to uncover more effective cancer treatments, help improve doctors’ understanding of rare diseases, and pave the way for more groundbreaking scientific discoveries in proteomics.
Protein sequencing is a long-standing challenge, widely regarded as one of biology's toughest problems. Unlike DNA, which consists of just four bases and follows a relatively simple sequencing method, proteins are composed of 20 amino acids arranged in endless combinations. Even small proteins can display staggering complexity.
Adding to the complexity, a protein's function can change entirely depending on how it folds into three-dimensional shapes. Many proteins also undergo chemical modifications after they are formed, which makes it incredibly difficult to trace these changes back to their original genetic blueprint.
To address these challenges, InstaNovo was developed as an AI-powered tool designed specifically for de novo protein sequencing. The term de novo refers to rebuilding protein sequences from scratch, rather than relying on existing reference databases.
In a paper published in Nature Machine Intelligence, the researchers reveal that InstaNovo is able to leverage AI to reconstruct peptide sequences from scratch, even for proteins that have not been analyzed before. Its breakthrough lies in its ability to decode fragmented peptide signals using a tailored deep-learning method delivering unprecedented efficiency and accuracy.
The InstaNovo+ model goes even further. It uses an iterative refinement process that aligns the peptide sequence more closely with spectral data. This is useful in the detection of chemically modified or hidden proteins.
The new AI tool is the result of a joint effort between InstaDeep, an AI company, and the Department of Biotechnology and Biomedicine at the Technical University of Denmark (DTU). Key contributors from DTU include Associate Professor Timothy Patrick Jenkins and Assistant Professor Konstantinos Kalogeropoulos.
The developers claim that the new tool could revolutionize protein sequencing, much like AlphaFold transformed protein structure prediction. In recognition of its impact, AlphaFold's creators were honored with the Nobel Prize in Chemistry in 2024 for their groundbreaking contributions to the field using AI.
“Together, our results and those of others show that scale is the most determining factor in de novo peptide sequencing model performance, as with other fields where the transformer architecture was employed," shared the researchers.
“We expect to further increase model performance by taking advantage of the vast amount of MS datasets available in repositories. We also anticipate widespread adoption by peers, and look forward to further exploration of fine-tuning, protein inference, and assembly, as well as building applications on top of our base model for hybrid or de novo searches.”
The creation of InstaNovo is not the first attempt by researchers to apply machine learning to protein sequencing. Earlier tools like the AI transformer protein decoder Casanovo showed how AI could help with protein sequencing, but they had a key limitation. Most of them depended heavily on reference databases, which made it hard to identify new or unique proteins.
The InstaNovo creators claim that their tool outperforms Casanovo in identifying peptide-spectrum matches (PSMs). The InstaNovo and InstaNovo+ identify 41.8% more PSMs than Casanovo, showcasing their superior capability in complex sequencing tasks.
“By eliminating dependency on protein databases and improving accuracy through iterative refinement, InstaNovo and InstaNovo+ uncover previously inaccessible proteomic landscapes, with the potential to drive discoveries across multiple scientific domains,” shared InstaDeep in a blog post.
The ability to go “database-free” is not just a side benefit, it is central to what makes InstaNovo innovative. However, the researchers admit that integrating the tool with existing laboratory workflows would be challenging. They also acknowledge that the outputs may require additional verification.
Nevertheless, the tools represent a significant step forward in protein research. With further refinement and more real-world testing, the tool can be useful in broadening our understanding of complex biological systems.



































































































































































![[The AI Show Episode 143]: ChatGPT Revenue Surge, New AGI Timelines, Amazon’s AI Agent, Claude for Education, Model Context Protocol & LLMs Pass the Turing Test](https://www.marketingaiinstitute.com/hubfs/ep%20143%20cover.png)

































































































![From Accountant to Data Engineer with Alyson La [Podcast #168]](https://cdn.hashnode.com/res/hashnode/image/upload/v1744420903260/fae4b593-d653-41eb-b70b-031591aa2f35.png?#)































































































.png?#)






































































































































![Apple Posts Full First Episode of 'Your Friends & Neighbors' on YouTube [Video]](https://www.iclarified.com/images/news/96990/96990/96990-640.jpg)

![Apple May Implement Global iPhone Price Increases to Mitigate Tariff Impacts [Report]](https://www.iclarified.com/images/news/96987/96987/96987-640.jpg)