IgCraft: AI Generates Human Antibodies for Drug Discovery & Engineering
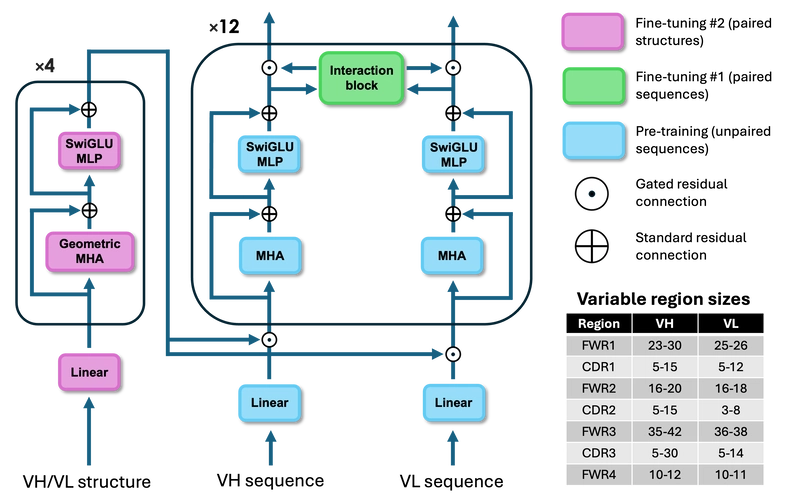
This is a Plain English Papers summary of a research paper called IgCraft: AI Generates Human Antibodies for Drug Discovery & Engineering. If you like these kinds of analysis, you should join AImodels.fyi or follow us on Twitter. Introducing IgCraft: A Swiss Army Knife for Antibody Design Monoclonal antibodies represent a growing segment of the pharmaceutical market, with their success as therapeutics stemming from both their binding capabilities and their favorable "developability" properties. Traditional antibody discovery methods involve either animal immunization or high-throughput library screening, each with their own tradeoffs. While in vitro screening offers faster, more ethical approaches, naturally-derived antibodies often exhibit better pharmacokinetics, specificity, and reduced immunogenicity. Machine learning has made significant progress in antibody design, but typically relies on separate task-specific models for sequence generation, structure-guided design, and other engineering tasks. This fragmentation creates workflow complexity that limits practical adoption. To address this challenge, researchers from the University of Cambridge have developed IgCraft, a unified framework that handles multiple antibody engineering tasks within a single model. IgCraft leverages Bayesian Flow Networks (BFNs) to generate paired human antibody sequences across diverse contexts. Similar to diffusion models but with key differences, BFNs perform joint denoising across all sequence positions, allowing flexible generation without imposing a specific order of tokens. This approach enables conditional generation of subsequences (inpainting) from a model trained only on full sequences, making it particularly suitable for the varied design scenarios encountered in antibody development. Unlike specialized approaches like AffinityFlow which focuses specifically on affinity maturation, IgCraft offers a comprehensive solution that can efficiently perform both unconditional and conditional sampling while incorporating structural information when available. How IgCraft Works: Technical Foundation Bayesian Flow Networks IgCraft's foundation rests on Bayesian Flow Networks (BFNs), a generative modeling approach for discrete tokens. Unlike diffusion models that denoise tokens directly, BFNs model a continuous denoising process over vectors of logits for different token categories. The BFN's generative process can be formulated as a stochastic differential equation (SDE) in logit space: For discrete tokens, a BFN approximates an SDE where a neural network predicts probabilities over token categories using relationships between noisy variables. IgCraft uses an accuracy schedule of α(t)=2t and implements a second-order solver to perform sampling in 20 steps. For conditional sampling, it employs a particle filtering method with 32 particles. The lack of a specific generation order in BFNs is particularly important, as it enables flexible sequence inpainting for arbitrary regions - a crucial capability given the multiple design scenarios encountered in antibody engineering. Network Architecture IgCraft employs a two-track transformer architecture specifically designed for paired antibody sequences: IgCraft's two-track transformer architecture. Layers are color-coded by the stage of training during which they are updated. The main backbone processes VH/VL sequences and outputs predicted amino acid probabilities at each position, with minimum/maximum lengths shown for each variable domain region. The architecture processes each antibody chain independently through separate transformer stacks with gated self-attention, rotary positional embeddings, pre-layer normalization, and SwiGLU transition layers. After each transformer block, token embeddings from both chains feed into interaction blocks using gated cross-attention, adaptive layer normalization, and conditional SwiGLU transition layers to integrate information between chains. For structural information encoding, IgCraft implements geometric multi-head attention from ESM3, with separate embedding layers for VH, VL, and epitope residues. This approach differs from Decoupled Sequence-Structure Generation, which separates sequence and structure generation into distinct processes. In total, IgCraft contains approximately 300M trainable parameters. Datasets and Training Regime IgCraft's training data combines paired and unpaired antibody sequences from multiple sources, with region-specific length constraints. The filtering process yielded training sets of 118M unpaired VH sequences, 135M unpaired VL sequences, and 1.5M paired VH/VL sequences. For structural data, the researchers clustered the training sequences, predicted structures using ABodyBuilder3-LM, and merged these with curated structures from SAbDab. The training process consists of three strategic stages: Pre-training each chain's transformer stack on
This is a Plain English Papers summary of a research paper called IgCraft: AI Generates Human Antibodies for Drug Discovery & Engineering. If you like these kinds of analysis, you should join AImodels.fyi or follow us on Twitter.
Introducing IgCraft: A Swiss Army Knife for Antibody Design
Monoclonal antibodies represent a growing segment of the pharmaceutical market, with their success as therapeutics stemming from both their binding capabilities and their favorable "developability" properties. Traditional antibody discovery methods involve either animal immunization or high-throughput library screening, each with their own tradeoffs. While in vitro screening offers faster, more ethical approaches, naturally-derived antibodies often exhibit better pharmacokinetics, specificity, and reduced immunogenicity.
Machine learning has made significant progress in antibody design, but typically relies on separate task-specific models for sequence generation, structure-guided design, and other engineering tasks. This fragmentation creates workflow complexity that limits practical adoption. To address this challenge, researchers from the University of Cambridge have developed IgCraft, a unified framework that handles multiple antibody engineering tasks within a single model.
IgCraft leverages Bayesian Flow Networks (BFNs) to generate paired human antibody sequences across diverse contexts. Similar to diffusion models but with key differences, BFNs perform joint denoising across all sequence positions, allowing flexible generation without imposing a specific order of tokens. This approach enables conditional generation of subsequences (inpainting) from a model trained only on full sequences, making it particularly suitable for the varied design scenarios encountered in antibody development.
Unlike specialized approaches like AffinityFlow which focuses specifically on affinity maturation, IgCraft offers a comprehensive solution that can efficiently perform both unconditional and conditional sampling while incorporating structural information when available.
How IgCraft Works: Technical Foundation
Bayesian Flow Networks
IgCraft's foundation rests on Bayesian Flow Networks (BFNs), a generative modeling approach for discrete tokens. Unlike diffusion models that denoise tokens directly, BFNs model a continuous denoising process over vectors of logits for different token categories. The BFN's generative process can be formulated as a stochastic differential equation (SDE) in logit space:
For discrete tokens, a BFN approximates an SDE where a neural network predicts probabilities over token categories using relationships between noisy variables. IgCraft uses an accuracy schedule of α(t)=2t and implements a second-order solver to perform sampling in 20 steps. For conditional sampling, it employs a particle filtering method with 32 particles.
The lack of a specific generation order in BFNs is particularly important, as it enables flexible sequence inpainting for arbitrary regions - a crucial capability given the multiple design scenarios encountered in antibody engineering.
Network Architecture
IgCraft employs a two-track transformer architecture specifically designed for paired antibody sequences:

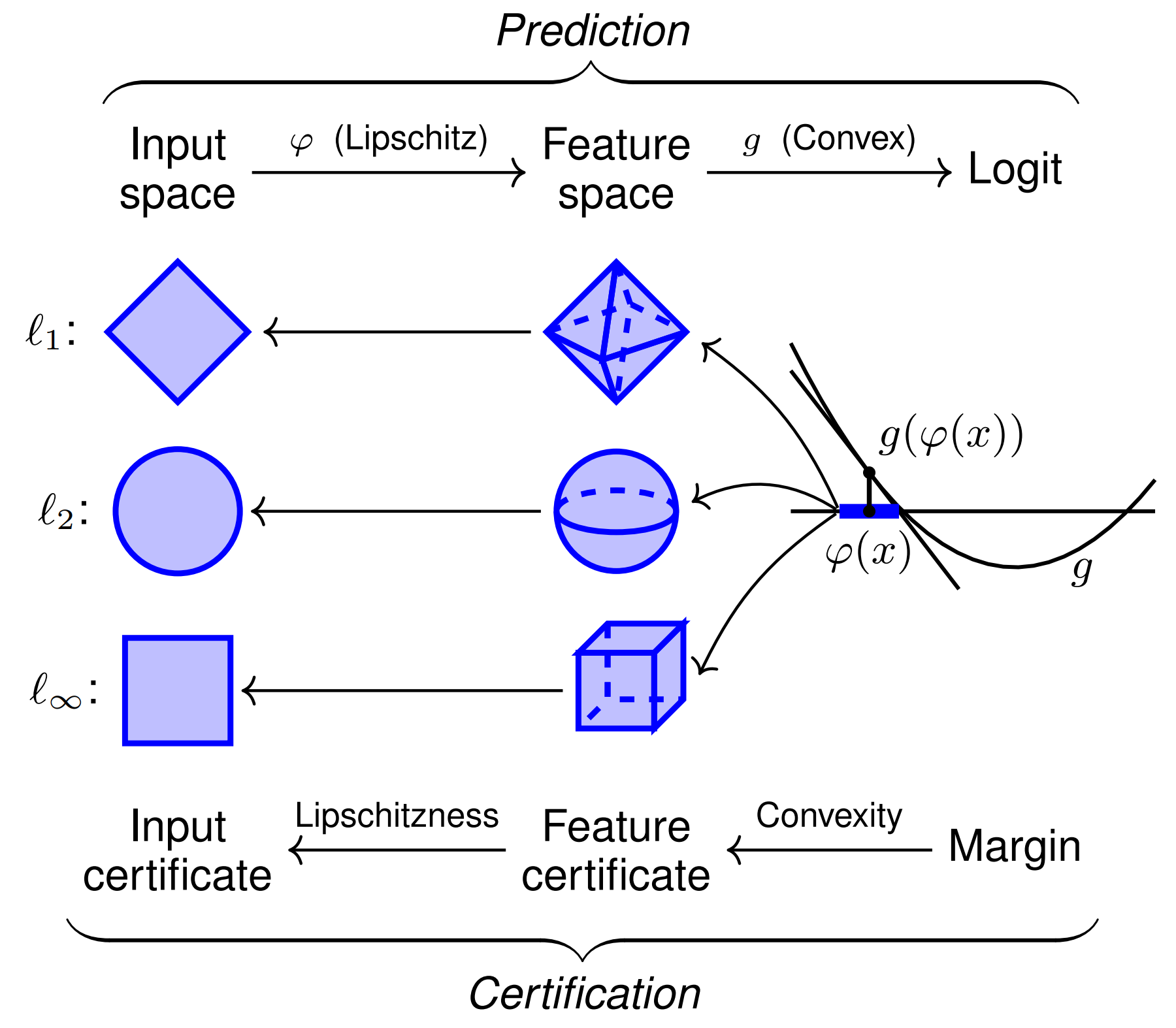
IgCraft's two-track transformer architecture. Layers are color-coded by the stage of training during which they are updated. The main backbone processes VH/VL sequences and outputs predicted amino acid probabilities at each position, with minimum/maximum lengths shown for each variable domain region.
The architecture processes each antibody chain independently through separate transformer stacks with gated self-attention, rotary positional embeddings, pre-layer normalization, and SwiGLU transition layers. After each transformer block, token embeddings from both chains feed into interaction blocks using gated cross-attention, adaptive layer normalization, and conditional SwiGLU transition layers to integrate information between chains.
For structural information encoding, IgCraft implements geometric multi-head attention from ESM3, with separate embedding layers for VH, VL, and epitope residues. This approach differs from Decoupled Sequence-Structure Generation, which separates sequence and structure generation into distinct processes. In total, IgCraft contains approximately 300M trainable parameters.
Datasets and Training Regime
IgCraft's training data combines paired and unpaired antibody sequences from multiple sources, with region-specific length constraints. The filtering process yielded training sets of 118M unpaired VH sequences, 135M unpaired VL sequences, and 1.5M paired VH/VL sequences. For structural data, the researchers clustered the training sequences, predicted structures using ABodyBuilder3-LM, and merged these with curated structures from SAbDab.
The training process consists of three strategic stages:
- Pre-training each chain's transformer stack on unpaired sequences
- Fine-tuning on paired sequences, with interaction blocks initially disabled
- Fine-tuning with paired antibody structures as conditioning information
This multi-stage approach ensures the model can function as both a pure sequence generator and incorporate structural information when available, similar to approaches that augment protein language models with structural adapters.
IgCraft in Action: Performance Across Multiple Tasks
Creating New Human-Like Antibody Sequences from Scratch
IgCraft demonstrates strong performance in generating novel, diverse, and realistic human antibody pairs. When compared against p-IgGen and real antibody sequences, IgCraft produces sequences with comparable characteristics:
| Samples | Novelty (VH / VL) | Diversity (VH / VL) | VH/VL mut. corr. |
|---|---|---|---|
| IgCraft | $10.5 / 2.7$ | $13.0 / 3.4$ | 0.49 |
| Reference | $10.2 / 2.7$ | $12.8 / 3.4$ | 0.55 |
| p-IgGen | $10.4 / 3.1$ | $12.7 / 3.7$ | 0.51 |
Table 1: Unconditional paired human antibody sampling results. Novelty shows each sequence's minimum edit distance to test set sequences, while diversity indicates minimum edit distance within the sample set. VH/VL pairing compatibility is estimated via correlation between germline sequence identities of heavy and light chains.
Additional metrics further confirm IgCraft's ability to generate sequences that closely match natural human antibodies:
| Samples | V seq. id. (%) | J seq. id. (%) | V diversity | J diversity | Humanness |
|---|---|---|---|---|---|
| IgCraft | 95.8 / 97.5 | 95.4 / 95.0 | 3.92 / 3.58 | 1.86 / 2.03 | 92.8 / 98.7 |
| Reference | 95.8 / 97.5 | 95.6 / 95.4 | 3.91 / 3.59 | 1.86 / 2.07 | 92.9 / 98.8 |
| p-IgGen | 96.1 / 97.0 | 95.0 / 95.1 | 3.91 / 3.67 | 1.91 / 2.09 | 93.0 / 98.7 |
Table 5: Additional unconditional sampling metrics showing germline sequence identity percentages, germline diversity (measured as shannon entropy), and humanness scores for both heavy and light chains.
These results demonstrate IgCraft's capability to generate antibody sequences that recapitulate the patterns of variation observed in natural human antibody repertoires, making it valuable for expanding the accessible design space in antibody discovery.
Filling in the Blanks: Sequence Inpainting Capabilities
One of IgCraft's key advantages is its ability to perform conditional sampling through sequence inpainting - generating missing regions while considering the surrounding context. When compared to AbLang2 and ESM3 on 2,000 test sequences, IgCraft shows competitive performance:
| Samples | H-CDR1 (%) | H-CDR2 (%) | H-CDR3 (%) | All CDRs (%) | All FWRs (%) |
|---|---|---|---|---|---|
| IgCraft | 91.6 | 89.4 | 41.1 | 74.2 | $\mathbf{96.7}$ |
| AbLang2 | $\mathbf{92.0}$ | $\mathbf{90.4}$ | $\mathbf{45.5}$ | $\mathbf{76.3}$ | 83.3 |
| ESM3-open | 77.7 | 58.4 | 34.1 | 54.1 | 13.0 |
Table 2: Mean amino acid recovery (AAR) for sequence inpainting on 2000 paired sequences. Results show recovery rates for individual heavy chain CDRs, all CDRs jointly, and all framework regions jointly.
IgCraft particularly excels at framework region inpainting (96.7% accuracy), significantly outperforming AbLang2 (83.3%) and ESM3 (13.0%) in this area. This capability is crucial for antibody humanization, where maintaining proper CDR-framework interactions is essential.
A more detailed breakdown of inpainting performance across variable regions shows consistent patterns:
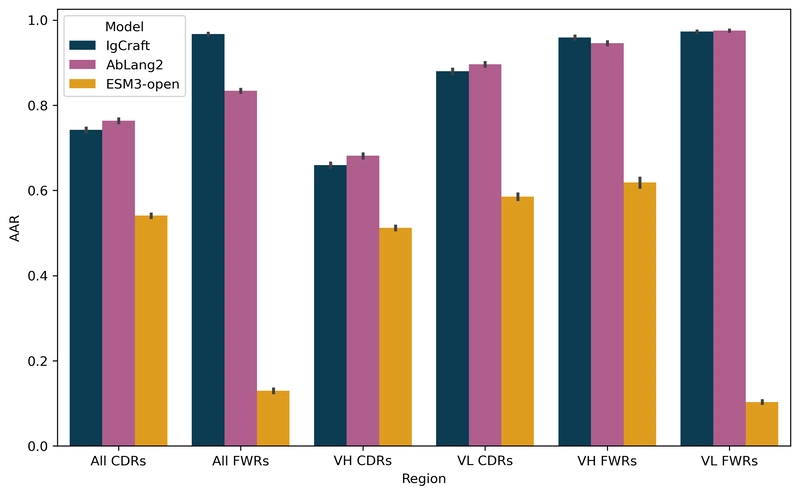
Mean amino acid recovery for sequence inpainting across different variable regions, showing IgCraft's relative strengths compared to AbLang2 and ESM3-open.
While Retrieval-Augmented Diffusion Models rely on retrieving relevant examples to guide inpainting, IgCraft accomplishes this task through its built-in conditional sampling capabilities, streamlining the workflow.
Designing Sequences from 3D Structures: Inverse Folding
IgCraft can also perform inverse folding - generating antibody sequences from 3D backbone structures. Testing on 98 human antibody structures against ProteinMPNN, AbMPNN, and Antifold revealed intriguing results:
| Samples | H-CDR1 (%) | H-CDR2 (%) | H-CDR3 (%) | FWR (%) | Humanness (VH / VL) |
Solubility (VH / VL) |
|---|---|---|---|---|---|---|
| IgCraft | 73.5 | 66.7 | 45.0 | 91.2 | $\mathbf{0.91} / \mathbf{0.97}$ | $\mathbf{0.08} / \mathbf{0.49}$ |
| Antifold | $\mathbf{77.1}$ | $\mathbf{75.3}$ | $\mathbf{58.1}$ | $\mathbf{92.7}$ | $0.89 / 0.95$ | $0.01 / 0.47$ |
| AbMPNN | 74.0 | 65.0 | 55.6 | 87.8 | $0.85 / 0.90$ | $0.02 / 0.42$ |
| ProteinMPNN | 48.9 | 46.4 | 31.9 | 58.9 | $0.51 / 0.51$ | $-0.12 / 0.11$ |
Table 3: Mean amino acid recovery and developability metrics for sequences generated by inverse folding models on 98 paired human antibody structures. IgCraft was the only method to improve both humanness and solubility metrics compared to the ground-truth antibodies.
While specialized antibody inverse folding tools achieved higher amino acid recovery rates, especially for H-CDR3, IgCraft significantly outperformed on humanness and solubility metrics. Notably, IgCraft was the only method to improve both metrics for both chains compared to the original sequences, highlighting its potential for structure-guided developability optimization.
Detailed per-region amino acid recovery statistics further illustrate these patterns:
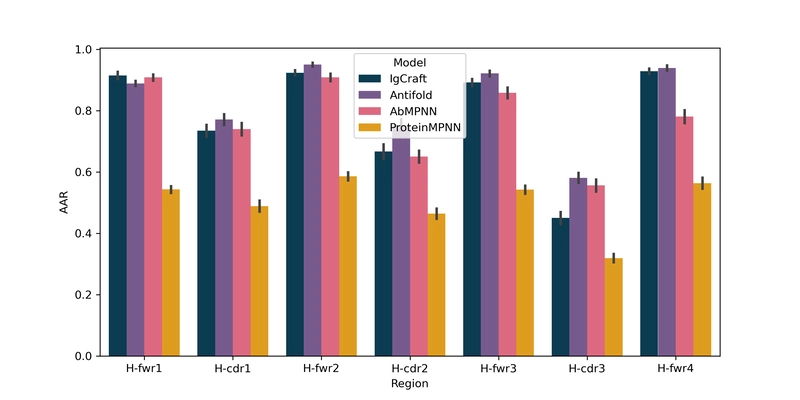
Amino acid recovery per-heavy chain region showing IgCraft's performance compared to specialized inverse folding models.
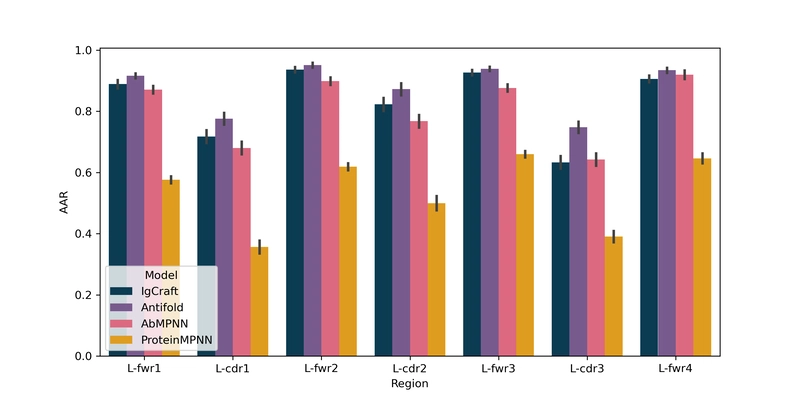
Amino acid recovery per-light chain variable region comparing IgCraft with other inverse folding approaches.
These results demonstrate IgCraft's ability to balance sequence recovery with improved developability properties, a crucial consideration for therapeutic antibody design.
Humanizing Mouse Antibodies: CDR Grafting Performance
"Humanizing" non-human antibodies remains a critical task in therapeutic development, as antibodies from other species often induce immune responses when administered to humans. IgCraft's conditional framework generation capabilities offer a powerful approach for CDR grafting - transferring mouse CDRs into human antibody frameworks.
When evaluated on 27 mouse antibodies against the specialized HuDiff model, IgCraft demonstrated impressive results:
| Samples | \% Seq. id. (VH / VL) |
Humanness (OASis) |
Humanness (AbNatiV, VH / VL) |
H-CDR3 RMSD ( $\AA$ ) | DockQ $>0.23$ |
|---|---|---|---|---|---|
| IgCraft | 77.6 / 77.5 | 77.9 | 0.88 / 0.90 | 2.04 | 10/20 |
| HuDiff | 81.4 / 80.3 | 74.6 | 0.87 / 0.82 | 1.99 | 9/20 |
| Reference | 100.0 / 100.0 | 47.3 | 0.68 / 0.64 | 1.87 | 11/20 |
Table 4: Evaluation of mouse CDR grafting on 27 paired mouse antibody structures from SAbDab. Results show sequence identity, humanness scores, structural fidelity (H-CDR3 RMSD), and binding functionality (DockQ >0.23).
IgCraft achieved better humanization than HuDiff according to the OASis metric (77.9 vs 74.6) and comparable AbNatiV scores, despite HuDiff being specifically fine-tuned to maximize this metric. Most importantly, IgCraft maintained key structural and functional properties, with similar H-CDR3 RMSD values and more correctly docked antibodies (10/20 vs 9/20), indicating better preservation of binding function.
Structural visualization confirms IgCraft's ability to preserve binding functionality while humanizing antibodies:

AlphaFold3 predictions showing how IgCraft's humanized antibodies maintain structural alignment with the original mouse antibodies. Examples show high-quality docking (left), acceptable docking (middle), and incorrect docking (right).
Unlike specialized humanization approaches like Generative Humanization, IgCraft performs this task as part of its broader unified framework while achieving competitive or superior results.
An ablation study confirmed the importance of structural information, showing that providing CDR structures as input significantly improved IgCraft's ability to preserve structural features during humanization:
| Samples | \% Seq. id. (VH / VL) |
Humanness (OASis) |
Humanness (AbNatiV, VH / VL) |
H-CDR3 RMSD ( $\overline{\mathbf{A}}$ ) | DockQ $>\mathbf{0 . 2 3}$ |
|---|---|---|---|---|---|
| Seq. only | $72.9 / 77.4$ | 78.4 | $0.85 / 0.91$ | 2.42 | $7 / 20$ |
| Seq. + structure | $77.6 / 77.5$ | 77.9 | $0.88 / 0.90$ | 2.04 | $10 / 20$ |
Table 6: Structural ablation study for IgCraft in CDR grafting, comparing sequence-only versus sequence+structure approaches.
Why IgCraft Matters: A Unified Approach to Antibody Engineering
IgCraft represents a significant advance in antibody engineering by providing the first generative model for paired antibody sequences capable of performing multiple design tasks through a unified architecture. Across unconditional generation, sequence inpainting, inverse folding, and CDR grafting, IgCraft achieves competitive or state-of-the-art performance while maintaining realistic, human-like sequence properties.
Particularly notable is IgCraft's exceptional performance in framework region inpainting and structure-based CDR grafting, where it achieves better humanization while maintaining functional features better than specialized approaches. Its ability to improve both humanness and solubility metrics during inverse folding demonstrates its potential for developability optimization.
The unified nature of IgCraft offers substantial practical advantages for antibody engineering workflows. Rather than using separate models for different tasks, researchers can employ a single tool for various design scenarios, from de novo generation to humanization to structure-guided optimization. This integration streamlines computational pipelines and enables more efficient exploration of the antibody design space.
As therapeutic antibodies continue to grow in importance within the pharmaceutical industry, tools like IgCraft that can balance binding affinity, developability, and manufacturability will become increasingly valuable. By demonstrating that a wide variety of antibody sequence generation tasks can be accomplished using a unified, scalable architecture, IgCraft establishes a promising foundation for the next generation of computational antibody design.


.jpg)
























![[Webinar] AI Is Already Inside Your SaaS Stack — Learn How to Prevent the Next Silent Breach](https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEiOWn65wd33dg2uO99NrtKbpYLfcepwOLidQDMls0HXKlA91k6HURluRA4WXgJRAZldEe1VReMQZyyYt1PgnoAn5JPpILsWlXIzmrBSs_TBoyPwO7hZrWouBg2-O3mdeoeSGY-l9_bsZB7vbpKjTSvG93zNytjxgTaMPqo9iq9Z5pGa05CJOs9uXpwHFT4/s1600/ai-cyber.jpg?#)






































































































































![[The AI Show Episode 144]: ChatGPT’s New Memory, Shopify CEO’s Leaked “AI First” Memo, Google Cloud Next Releases, o3 and o4-mini Coming Soon & Llama 4’s Rocky Launch](https://www.marketingaiinstitute.com/hubfs/ep%20144%20cover.png)










































































































































































![Rogue Company Elite tier list of best characters [April 2025]](https://media.pocketgamer.com/artwork/na-33136-1657102075/rogue-company-ios-android-tier-cover.jpg?#)






































































_Andreas_Prott_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)






















































































![What’s new in Android’s April 2025 Google System Updates [U: 4/18]](https://i0.wp.com/9to5google.com/wp-content/uploads/sites/4/2025/01/google-play-services-3.jpg?resize=1200%2C628&quality=82&strip=all&ssl=1)










![Apple Watch Series 10 Back On Sale for $299! [Lowest Price Ever]](https://www.iclarified.com/images/news/96657/96657/96657-640.jpg)
![EU Postpones Apple App Store Fines Amid Tariff Negotiations [Report]](https://www.iclarified.com/images/news/97068/97068/97068-640.jpg)
![Apple Slips to Fifth in China's Smartphone Market with 9% Decline [Report]](https://www.iclarified.com/images/news/97065/97065/97065-640.jpg)