From Scripts to Cloud: My Hands-On Guide to ML + DevOps
As a DevOps engineer, I'm used to automating backend systems, deploying apps, and scaling infrastructure. But when I trained my first machine learning model, I found myself asking: How do I bring the same level of automation and structure here? I also started wondering, as AI continues to evolve, what’s the fate of a Cloud/DevOps engineer? Or better still, what role will we play in this new landscape? This blog post is my attempt to explore and answer those questions by integrating AI and DevOps. Think of it as a beginner-friendly hands-on project guide to building a bridge between the two worlds. What I Built To put these questions into practice, I built a simple yet meaningful project: A machine learning model that predicts epitope regions in a protein sequence. An epitope is the part of an antigen that an antibody binds to, helping the immune system recognize and fight infections. This makes epitope prediction really important in vaccine and drug development. To bring DevOps into the mix, I wrapped the model in a FastAPI backend, containerized it with Docker, and set up automated deployment to an EC2 instance using GitHub Actions. Why This Matters This ML model helps researchers, especially in the low resource environments by accelerating their work, cut costs, and reduce the time spent running multiple experiments to identify which part of a protein is epitopic. By integrating DevOps practices, this model is accessible even to researchers without a strong data science or technical background. All they need to do is input their protein sequences into the cloud-hosted web UI and the prediction is returned instantly. This doesn’t just speed up research, it also widens access and promotes inclusion in the global scientific space. Tools Used & Installed Python 3.8 and above FastAPI Conda Git GitHub Actions Docker AWS EC2 Prerequisite Knowledge To get the most out of this blog, it helps to be familiar with: Linux command-line basics Python programming Basic machine learning concepts Version control with Git Fundamentals of Docker Understanding of APIs (e.g FastAPI) Basic CI/CD workflows Conda (for managing Python environments) Project Overview The project is structured to separate concerns clearly: Data Scripts: For retrieving, preprocessing, and training the model Notebook: Used for exploratory data analysis and visualization Model Artifacts: Trained model saved for inference API Backend: A FastAPI app that serves predictions Dockerfile: Containerizes the app for deployment CI/CD Workflow: GitHub Actions automates the build and deployment Cloud Infrastructure: The app is deployed to an EC2 instance on AWS The full codebase and directory structure can be found in my github repository Build With Me Let’s walk through how I brought everything together, from training the ML model to deploying it in the cloud. Setting up the environment I created a Conda environment and installed all necessary dependencies: conda create -n epitope python=3.10 conda activate epitope pip install -r requirements.txt To build with me, clone the repository git clone https://github.com/AzeematRaji/epitope-ml-model.git cd epitope-ml-model Training the Machine Learning Model I retrieved and preprocessed the data: python scripts/retrieve.py python scripts/preprocess.py Then I trained and saved the model: python scripts/train.py joblib.dump(model, "../models/epitope_model.joblib") Creating the FastAPI Backend I built a simple API using FastAPI to serve the trained model. To test it locally: uvicorn app.main:app --host 0.0.0.0 --port 8000 --reload Test access via API from the Browser: localhost:8000 Writing a Dockerfile I containerized the app using Docker: docker build -t epitope-api . docker run -p 8000:8000 epitope-api This made it easy to run in any environment. Test access via API from the Browser: localhost:8000 Now, we have confirmed the app is working locally. Lets move it to the cloud. Setting Up the EC2 Instance Launch an EC2 Instance on AWS Choose an appropriate instance type (e.g., t3.medium because of data size). Configure the security group to allow inbound traffic on port 8000 from 0.0.0.0 for testing. Creating the GitHub Actions workflow To automate deployment, I created a GitHub Actions workflow .github/workflows/deploy.yml that is triggered whenever there is a push to the main branch, it: - Containerizes the application, - Pushes it to DockerHub, - Deploys it on your EC2 instance. GitHub Secrets Configuration In the GitHub repository, the following secrets need to be configured to ensure the pipeline works: DOCKER_USERNAME DOCKER_PASSWORD EC2_SSH_KEY: The private SSH key or .pem file used to connect to the EC2 instance EC2_PUBLIC_IP This process eliminates the need to manually deploy the application every time there is a code change, streamlining t
As a DevOps engineer, I'm used to automating backend systems, deploying apps, and scaling infrastructure. But when I trained my first machine learning model, I found myself asking:
How do I bring the same level of automation and structure here?
I also started wondering, as AI continues to evolve, what’s the fate of a Cloud/DevOps engineer? Or better still, what role will we play in this new landscape?
This blog post is my attempt to explore and answer those questions by integrating AI and DevOps. Think of it as a beginner-friendly hands-on project guide to building a bridge between the two worlds.
What I Built
To put these questions into practice, I built a simple yet meaningful project:
A machine learning model that predicts epitope regions in a protein sequence.
An epitope is the part of an antigen that an antibody binds to, helping the immune system recognize and fight infections. This makes epitope prediction really important in vaccine and drug development.
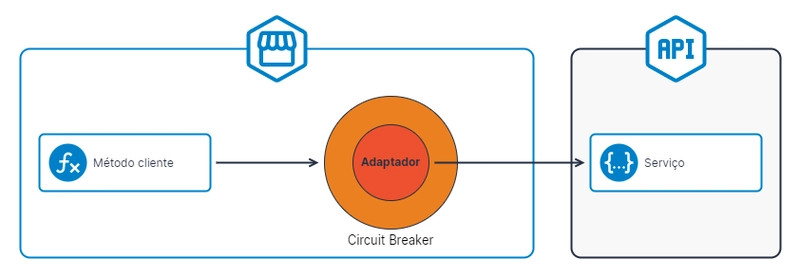
To bring DevOps into the mix, I wrapped the model in a FastAPI backend, containerized it with Docker, and set up automated deployment to an EC2 instance using GitHub Actions.
Why This Matters
This ML model helps researchers, especially in the low resource environments by accelerating their work, cut costs, and reduce the time spent running multiple experiments to identify which part of a protein is epitopic.
By integrating DevOps practices, this model is accessible even to researchers without a strong data science or technical background. All they need to do is input their protein sequences into the cloud-hosted web UI and the prediction is returned instantly.
This doesn’t just speed up research, it also widens access and promotes inclusion in the global scientific space.
Tools Used & Installed
- Python 3.8 and above
- FastAPI
- Conda
- Git
- GitHub Actions
- Docker
- AWS EC2
Prerequisite Knowledge
To get the most out of this blog, it helps to be familiar with:
- Linux command-line basics
- Python programming
- Basic machine learning concepts
- Version control with Git
- Fundamentals of Docker
- Understanding of APIs (e.g FastAPI)
- Basic CI/CD workflows
- Conda (for managing Python environments)
Project Overview
The project is structured to separate concerns clearly:
- Data Scripts: For retrieving, preprocessing, and training the model
- Notebook: Used for exploratory data analysis and visualization
- Model Artifacts: Trained model saved for inference
- API Backend: A FastAPI app that serves predictions
- Dockerfile: Containerizes the app for deployment
- CI/CD Workflow: GitHub Actions automates the build and deployment
- Cloud Infrastructure: The app is deployed to an EC2 instance on AWS
The full codebase and directory structure can be found in my github repository
Build With Me
Let’s walk through how I brought everything together, from training the ML model to deploying it in the cloud.
- Setting up the environment
I created a Conda environment and installed all necessary dependencies:
conda create -n epitope python=3.10
conda activate epitope
pip install -r requirements.txt
- To build with me, clone the repository
git clone https://github.com/AzeematRaji/epitope-ml-model.git
cd epitope-ml-model
- Training the Machine Learning Model
I retrieved and preprocessed the data:
python scripts/retrieve.py
python scripts/preprocess.py
Then I trained and saved the model:
python scripts/train.py
joblib.dump(model, "../models/epitope_model.joblib")
- Creating the FastAPI Backend I built a simple API using FastAPI to serve the trained model. To test it locally:
uvicorn app.main:app --host 0.0.0.0 --port 8000 --reload
Test access via API from the Browser:
localhost:8000
- Writing a Dockerfile
I containerized the app using Docker:
docker build -t epitope-api .
docker run -p 8000:8000 epitope-api
This made it easy to run in any environment.
Test access via API from the Browser:
localhost:8000
Now, we have confirmed the app is working locally. Lets move it to the cloud.
-
Setting Up the EC2 Instance
- Launch an EC2 Instance on AWS
- Choose an appropriate instance type (e.g., t3.medium because of data size).
- Configure the security group to allow inbound traffic on port 8000 from 0.0.0.0 for testing.
Creating the GitHub Actions workflow
To automate deployment, I created a GitHub Actions workflow .github/workflows/deploy.yml that is triggered whenever there is a push to the main branch, it:
- Containerizes the application,
- Pushes it to DockerHub,
- Deploys it on your EC2 instance.
- GitHub Secrets Configuration
In the GitHub repository, the following secrets need to be configured to ensure the pipeline works:
DOCKER_USERNAME
DOCKER_PASSWORD
EC2_SSH_KEY: The private SSH key or .pem file used to connect to the EC2 instance
EC2_PUBLIC_IP
This process eliminates the need to manually deploy the application every time there is a code change, streamlining the workflow.
- Final testing and accessibility
Access the FastAPI via the browser:
http://
This would display a UI prompt to input protein sequence and predict whether it epitope or non-epitope
Lessons Learnt
After evaluating the model, I observed that it tends to favor the prediction of non-epitope sequences over epitope sequences. This bias is largely due to the highly imbalanced dataset used during training.
This highlighted two important takeaways:
Data quality matters: An imbalanced dataset can significantly affect a model's performance, especially in critical applications like drug discovery.
There's room for improvement: Future iterations could involve experimenting with different featurizers, applying sampling techniques (e.g. SMOTE), and trying out more balanced or larger datasets to improve accuracy and fairness in predictions
Raw Reflections & What’s Ahead
This project was just the beginning of my exploration into the intersection of AI and DevOps. While I kept things simple here, there’s so much more to integrate, from using Kubernetes to manage and scale more complex ML workloads, to Terraform for automating infrastructure provisioning in a more structured and repeatable way.
These tools and ideas will be explored in future projects and blog posts, where I’ll continue building and documenting how DevOps can enhance the accessibility and reliability of AI applications.
If you found this insightful, feel free to follow my journey on GitHub where I share more projects like this.


































































































































































![[The AI Show Episode 144]: ChatGPT’s New Memory, Shopify CEO’s Leaked “AI First” Memo, Google Cloud Next Releases, o3 and o4-mini Coming Soon & Llama 4’s Rocky Launch](https://www.marketingaiinstitute.com/hubfs/ep%20144%20cover.png)














































































































![From fast food worker to cybersecurity engineer with Tae'lur Alexis [Podcast #169]](https://cdn.hashnode.com/res/hashnode/image/upload/v1745242807605/8a6cf71c-144f-4c91-9532-62d7c92c0f65.png?#)




















![BPMN-procesmodellering [closed]](https://i.sstatic.net/l7l8q49F.png)















































































.jpg?#)
.jpg?#)


























































































































![CarPlay app with web browser for streaming video hits App Store [U]](https://i0.wp.com/9to5mac.com/wp-content/uploads/sites/6/2024/11/carplay-apple.jpeg?resize=1200%2C628&quality=82&strip=all&ssl=1)

![What’s new in Android’s April 2025 Google System Updates [U: 4/21]](https://i0.wp.com/9to5google.com/wp-content/uploads/sites/4/2025/01/google-play-services-3.jpg?resize=1200%2C628&quality=82&strip=all&ssl=1)












![Apple Releases iOS 18.5 Beta 3 and iPadOS 18.5 Beta 3 [Download]](https://www.iclarified.com/images/news/97076/97076/97076-640.jpg)
![Apple Seeds visionOS 2.5 Beta 3 to Developers [Download]](https://www.iclarified.com/images/news/97077/97077/97077-640.jpg)
![Apple Seeds tvOS 18.5 Beta 3 to Developers [Download]](https://www.iclarified.com/images/news/97078/97078/97078-640.jpg)
![Apple Seeds watchOS 11.5 Beta 3 to Developers [Download]](https://www.iclarified.com/images/news/97079/97079/97079-640.jpg)