DIABETES PREDICTION APP WITH MACHINE LEARNING
PROJECT INTRODUCTION Diabetes is a condition that affects how your body processes sugar (glucose). Typically, your body uses insulin to help regulate blood sugar levels, but in diabetes, this process gets disrupted. There are two main types: Type 1 Diabetes: The body doesn’t produce insulin at all. It usually develops early in life and requires insulin injections. Type 2 Diabetes: occurs when the body either doesn’t produce enough insulin or can’t use it properly. It’s more common and often linked to lifestyle factors like diet and exercise. If left unmanaged, diabetes can lead to serious health problems, but with the right care, like a balanced diet, exercise, and medication, it can be controlled. That’s where your Diabetes Prediction App comes in, helping people get an early indication and take action! PROJECT AIM The dataset for this project was downloaded from Kaggle. This project aims to develop an app that can predict whether a patient is diabetic. Data handling and visualization will also take place to gain insight. A Logistic Regression and Random Forest classifier model would be created, and the best-performing model would be used to determine if a patient is diabetic or not. DATASET OVERVIEW The dataset is obtained from https://www.kaggle.com/datasets/uciml/pima-indians-diabetes-database Import Required Libraries #Import required libraries import pandas as pd import numpy as np import matplotlib.pyplot as plt import seaborn as sns import warnings warnings.filterwarnings('ignore') %matplotlib inline sns.set_style('whitegrid') Load The Dataset df = pd.read_csv('diabetes.csv') df.head(5) Get Dataset Information #information of dataset df.info() Information about dataset attributes Pregnancies: To express the Number of pregnancies Glucose: To express the Glucose level in blood BloodPressure: To express the Blood pressure measurement SkinThickness: To express the thickness of the skin Insulin: To express the Insulin level in blood BMI: To express the Body mass index DiabetesPedigreeFunction: To express the Diabetes percentage Age: To express the age Outcome: To express the final result, 1 is Yes and 0 is No Dataset Statistics #check statistics of dataset df.describe().T Observation: The dataset's statistics show that the minimum values of Glucose, Blood Pressure, Skin Thickness, Insulin, and BMI cannot realistically be 0, so this is a case that must be dealt with. Data Handling We would check for missing values in this aspect and handle them accordingly. #Check for missing values df.isna().sum() Observation: No missing values in the dataset. Handling Zero Values In this aspect, we would handle the zeros in the dataset. Firstly, we would check where the zero appears. #check for where 0 is present in each column print(df[df['Glucose'] == 0].shape[0]) print(df[df['BloodPressure'] == 0].shape[0]) print(df[df['SkinThickness'] == 0].shape[0]) print(df[df['Insulin'] == 0].shape[0]) print(df[df['BMI'] == 0].shape[0]) Output: 5 35 227 374 11 Next, we would visualize the plot to check the distribution of each column. #Check distribution of each column in the dataset df.hist(figsize=(20,20)) plt.show() Observation: Some of the columns have skewed distributions, so the mean is less affected by outliers than the median. Glucose and Blood Pressure have normal distributions; hence, we replace 0 values in those columns with mean values. Skin thickness, Insulin, and BMI have skewed distributions; hence, the median is a better choice as it is less affected by outliers. #Handling Zero Values df['Glucose'] = df['Glucose'].replace(0, df['Glucose'].mean()) df['BloodPressure'] = df['BloodPressure'].replace(0, df['BloodPressure'].mean()) df['SkinThickness'] = df['SkinThickness'].replace(0, df['SkinThickness'].median()) df['Insulin'] = df['Insulin'].replace(0, df['Insulin'].median()) df['BMI'] = df['BMI'].replace(0, df['BMI'].median()) Data Visualizations In this aspect, we would perform a simple visualization where we check the relationship between the target column(Outcome) with the other columns. #Get numerical columns num_col = ['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age'] #Visualize columns in respect to the outcome. # Number of rows needed (assuming you want 2 histograms per row) nrows = (len(num_col) + 1) // 2 # this will round up the division fig, axes = plt.subplots(nrows=nrows, ncols=2, figsize=(10, nrows * 5)) # Flatten axes array to make it easier to iterate over axes = axes.flatten() for i, col in enumerate(num_col): sns.histplot(df, x=col, hue=df['Outcome'], ax=axes[i]) axes[i].set_title(f'Distribution of {col} by Outcome') # Hide any unused subplots if there are an odd number of columns for j in range(i + 1, len(axes)): axes[j].axis('off') plt.tig

PROJECT INTRODUCTION
Diabetes is a condition that affects how your body processes sugar (glucose). Typically, your body uses insulin to help regulate blood sugar levels, but in diabetes, this process gets disrupted. There are two main types:
- Type 1 Diabetes: The body doesn’t produce insulin at all. It usually develops early in life and requires insulin injections.
- Type 2 Diabetes: occurs when the body either doesn’t produce enough insulin or can’t use it properly. It’s more common and often linked to lifestyle factors like diet and exercise.
If left unmanaged, diabetes can lead to serious health problems, but with the right care, like a balanced diet, exercise, and medication, it can be controlled. That’s where your Diabetes Prediction App comes in, helping people get an early indication and take action!
PROJECT AIM
The dataset for this project was downloaded from Kaggle. This project aims to develop an app that can predict whether a patient is diabetic. Data handling and visualization will also take place to gain insight. A Logistic Regression and Random Forest classifier model would be created, and the best-performing model would be used to determine if a patient is diabetic or not.
DATASET OVERVIEW
The dataset is obtained from https://www.kaggle.com/datasets/uciml/pima-indians-diabetes-database
Import Required Libraries
#Import required libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
sns.set_style('whitegrid')
Load The Dataset
df = pd.read_csv('diabetes.csv')
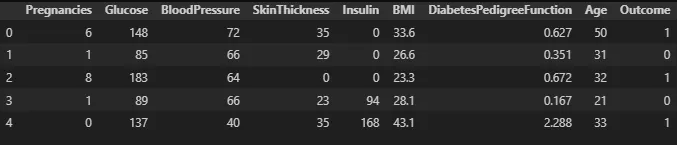
df.head(5)
Get Dataset Information
#information of dataset
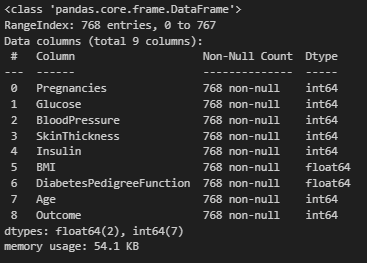
df.info()
Information about dataset attributes
- Pregnancies: To express the Number of pregnancies
- Glucose: To express the Glucose level in blood
- BloodPressure: To express the Blood pressure measurement
- SkinThickness: To express the thickness of the skin
- Insulin: To express the Insulin level in blood
- BMI: To express the Body mass index
- DiabetesPedigreeFunction: To express the Diabetes percentage
- Age: To express the age
- Outcome: To express the final result, 1 is Yes and 0 is No
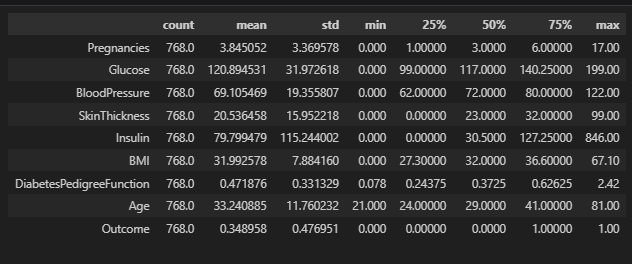
Dataset Statistics
#check statistics of dataset
df.describe().T
Observation:
- The dataset's statistics show that the minimum values of Glucose, Blood Pressure, Skin Thickness, Insulin, and BMI cannot realistically be 0, so this is a case that must be dealt with.
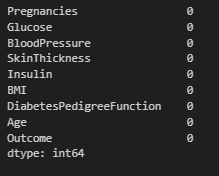
Data Handling
We would check for missing values in this aspect and handle them accordingly.
#Check for missing values
df.isna().sum()
Observation:
- No missing values in the dataset.
Handling Zero Values
In this aspect, we would handle the zeros in the dataset.
Firstly, we would check where the zero appears.
#check for where 0 is present in each column
print(df[df['Glucose'] == 0].shape[0])
print(df[df['BloodPressure'] == 0].shape[0])
print(df[df['SkinThickness'] == 0].shape[0])
print(df[df['Insulin'] == 0].shape[0])
print(df[df['BMI'] == 0].shape[0])
Output:
5
35
227
374
11
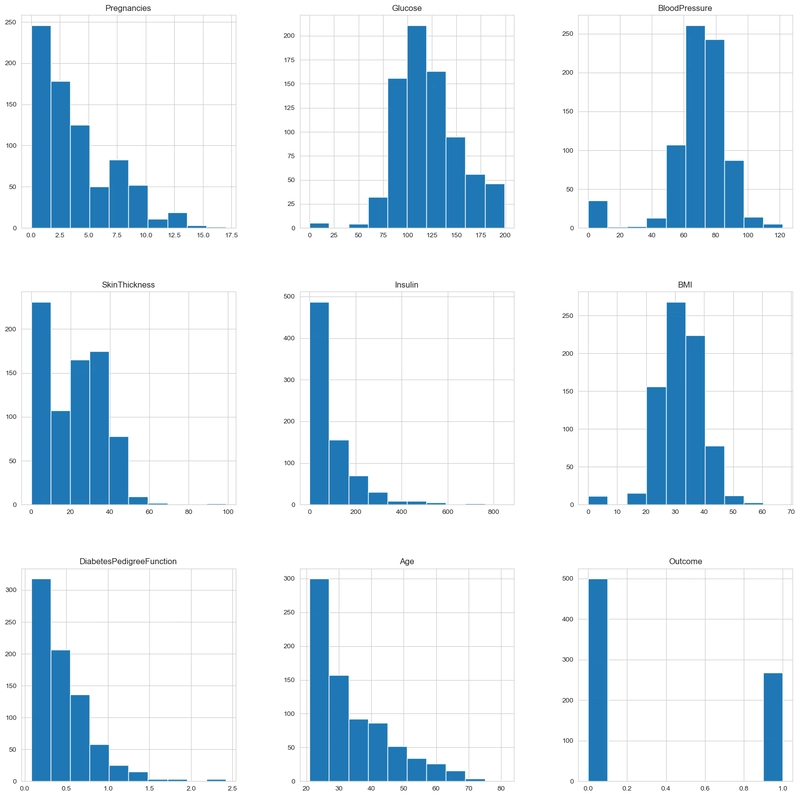
Next, we would visualize the plot to check the distribution of each column.
#Check distribution of each column in the dataset
df.hist(figsize=(20,20))
plt.show()
Observation:
- Some of the columns have skewed distributions, so the mean is less affected by outliers than the median. Glucose and Blood Pressure have normal distributions; hence, we replace 0 values in those columns with mean values. Skin thickness, Insulin, and BMI have skewed distributions; hence, the median is a better choice as it is less affected by outliers.
#Handling Zero Values
df['Glucose'] = df['Glucose'].replace(0, df['Glucose'].mean())
df['BloodPressure'] = df['BloodPressure'].replace(0, df['BloodPressure'].mean())
df['SkinThickness'] = df['SkinThickness'].replace(0, df['SkinThickness'].median())
df['Insulin'] = df['Insulin'].replace(0, df['Insulin'].median())
df['BMI'] = df['BMI'].replace(0, df['BMI'].median())
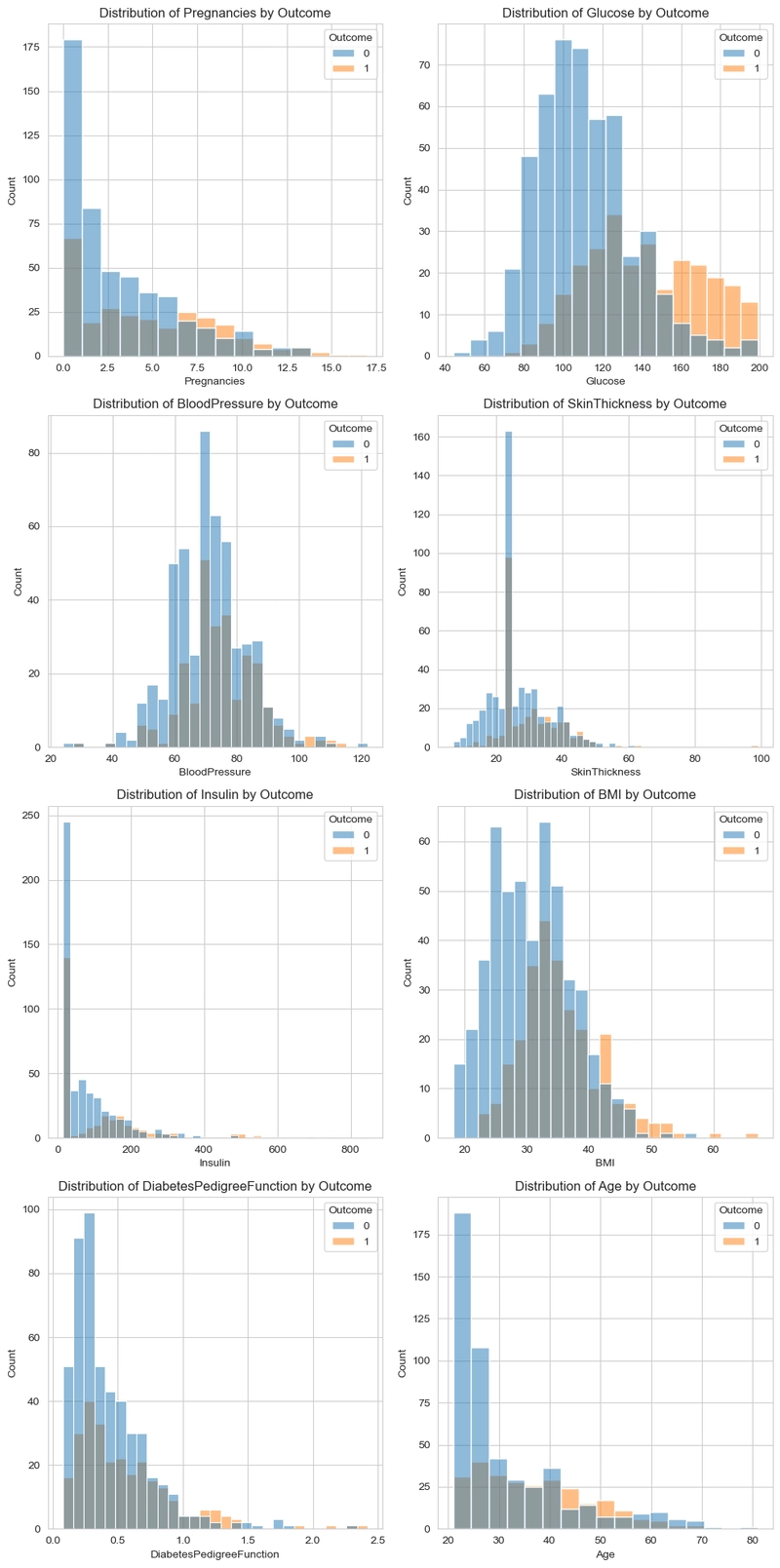
Data Visualizations
In this aspect, we would perform a simple visualization where we check the relationship between the target column(Outcome) with the other columns.
#Get numerical columns
num_col = ['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin',
'BMI', 'DiabetesPedigreeFunction', 'Age']
#Visualize columns in respect to the outcome.
# Number of rows needed (assuming you want 2 histograms per row)
nrows = (len(num_col) + 1) // 2 # this will round up the division
fig, axes = plt.subplots(nrows=nrows, ncols=2, figsize=(10, nrows * 5))
# Flatten axes array to make it easier to iterate over
axes = axes.flatten()
for i, col in enumerate(num_col):
sns.histplot(df, x=col, hue=df['Outcome'], ax=axes[i])
axes[i].set_title(f'Distribution of {col} by Outcome')
# Hide any unused subplots if there are an odd number of columns
for j in range(i + 1, len(axes)):
axes[j].axis('off')
plt.tight_layout()
plt.show()
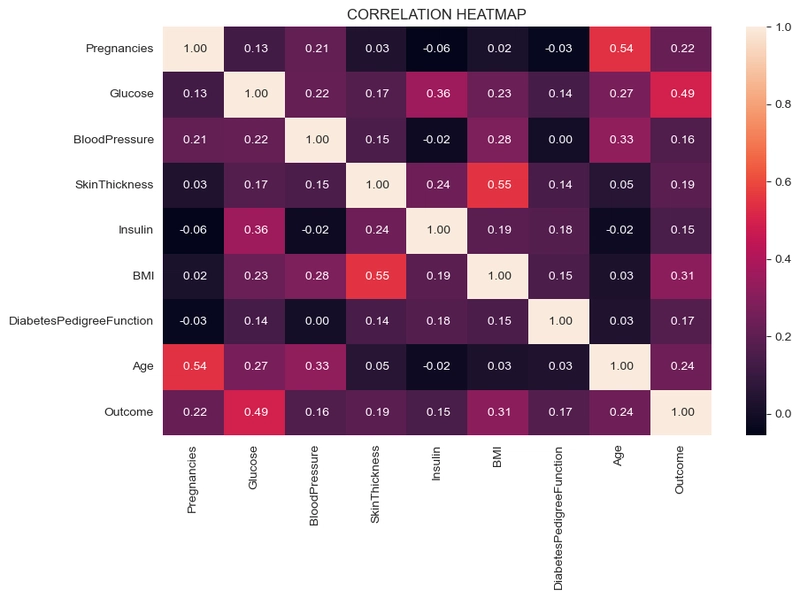
Correlation Heatmap
#correlation heatmap
plt.figure(figsize=(10,6))
sns.heatmap(df.corr(), annot=True, fmt=' .2f')
plt.title('CORRELATION HEATMAP')
plt.show()
Data Preparation
In this aspect, I would first scale the dataset using the standard scaler and split into X(Feature variable) and y(Target variable).
#Scale data
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X = pd.DataFrame(scaler.fit_transform(df.drop(columns=['Outcome'])), columns=df.columns[:-1])
y = df['Outcome']
y
Then, I would split the dataset into train and test splits using the scikit-learn TrainTestSplit.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)
Observation:
- The dataset was split into Features [X] and Target[y] variable
- It was then split into our Train and Test splits using TestTrainSplit.
- The dataset was split into 80% train data and 20% test data.
Model Selection and Evaluation
We used two models for this prediction project, models used are:
- Logistic Regression: a statistical method used to predict the probability of a binary outcome (like yes/no, 0/1) based on one or more independent variables, essentially predicting the likelihood of an event occurring.
- Random Forest Classifier: a machine learning algorithm that uses an ensemble of decision trees to classify data, making predictions by averaging the predictions of individual trees. It’s a powerful and versatile tool known for its accuracy and efficiency.
Logistic Regression
Build the model for prediction.
#import required libraries
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, classification_report, confusion_matrix
lr = LogisticRegression()
lr.fit(X_train,y_train)
#predictions
train_pred = lr.predict(X_train) #prediction on training set
test_pred = lr.predict(X_test) #Prediction on test set
#Accuracy scores
train_acc = accuracy_score(y_train,train_pred)
test_acc = accuracy_score(y_test, test_pred)
print('Train Set Accuracy: ', train_acc * 100)
print('Test Set Accuracy: ', test_acc * 100)
print()
#Confusion matrix and classification report
print('Confusion Matrix:\n', confusion_matrix(y_test,test_pred))
print('Classification Report:\n', classification_report(y_test,test_pred))
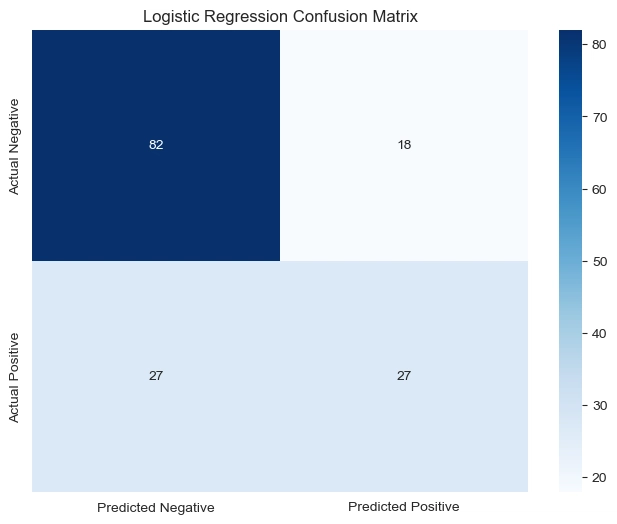
#Visualize the Logistic Regression confusion matrix
#convert to matrix
conf_matrix = np.array([[82, 18], [27,27]])
#convert to dataframe
df_cm = pd.DataFrame(conf_matrix, index=['Actual Negative', 'Actual Positive'], columns=['Predicted Negative', 'Predicted Positive'])
#heatmap
plt.figure(figsize=(8,6))
sns.heatmap(df_cm, annot=True, fmt='d', cmap='Blues')
plt.title('Logistic Regression Confusion Matrix')
plt.show()
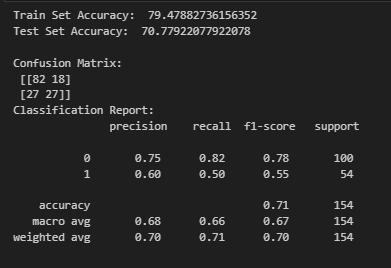
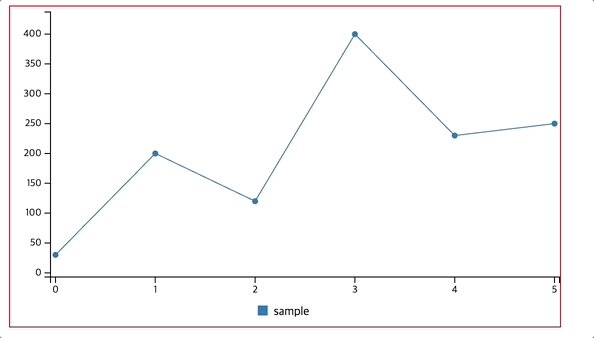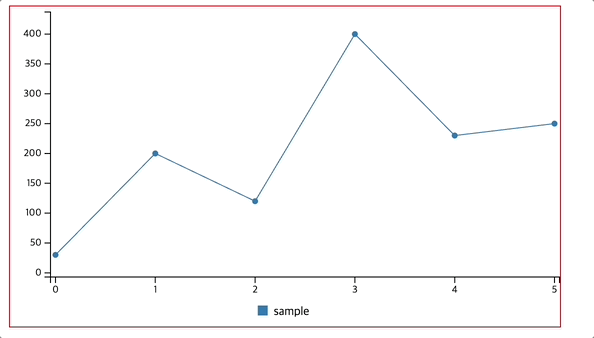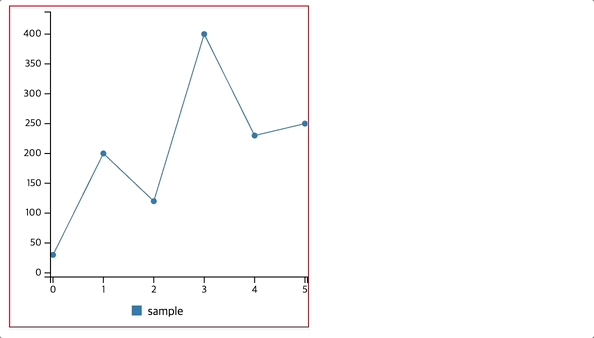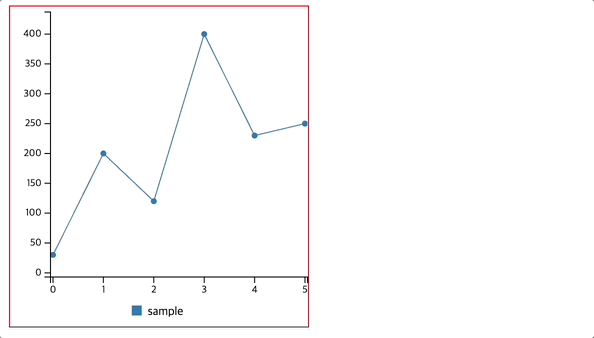
Observation:
The model achieves 79.48% accuracy on the training set and 70.78% accuracy on the test set, indicating a moderate drop in performance, which suggests some overfitting.
From the confusion matrix, the model correctly classifies 82 non-diabetic patients but misclassifies 18 as diabetic. It also correctly classifies 27 diabetic patients but misclassifies 27 as non-diabetic, which may indicate difficulty in distinguishing diabetic cases.
The classification report shows that the model has higher precision (0.75) and recall (0.82) for non-diabetic cases compared to diabetic cases (precision = 0.60, recall = 0.50). This suggests that the model is better at identifying non-diabetic patients but struggles with diabetic cases, likely due to class imbalance or feature representation.
Random Forest Classifier
Build the model for prediction.
#import required libraries
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, classification_report, confusion_matrix
from sklearn.model_selection import GridSearchCV
#hyperparameter grid
param_grid = {
'n_estimators': [50, 100, 200, 300],
'max_depth': [10, 20 ,30],
'min_samples_split': [2, 5, 10]
}
#Perform gridsearch with cross validation
grid = GridSearchCV(RandomForestClassifier(), param_grid, cv=5, scoring='accuracy', n_jobs=-1)
grid.fit(X_train,y_train)
#get the best estimator
print('Best param: ', grid.best_params_)
rfc = grid.best_estimator_
#predictions
rf_train_pred = rfc.predict(X_train)
rf_test_pred = rfc.predict(X_test)
#Accuracy score
rf_train_acc = accuracy_score(y_train,rf_train_pred)
rf_test_acc = accuracy_score(y_test, rf_test_pred)
print('Train Set Accuracy: ', rf_train_acc * 100)
print('Test Set Accuracy: ', rf_test_acc * 100)
print()
#Confusion matrix and classification report
print('Confusion Matrix:\n', confusion_matrix(y_test,rf_test_pred))
print('Classification Report:\n', classification_report(y_test,rf_test_pred))
#visualize the confusion matrix
#convert to matrix
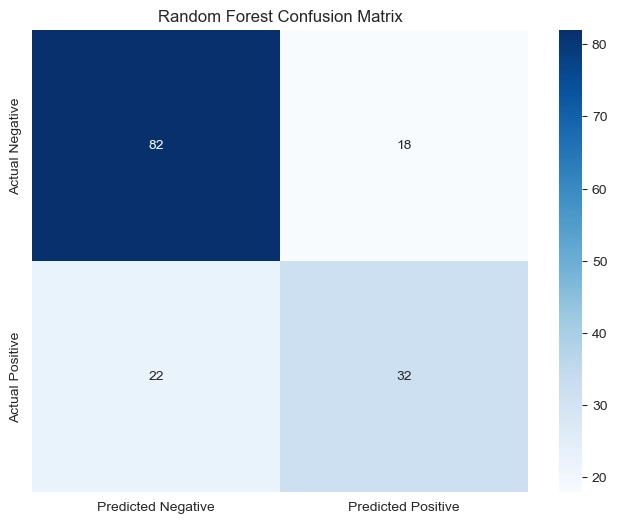
rf_matrix = np.array([[82,18],[22,32]])
#convert to dataframe
rf_df = pd.DataFrame(rf_matrix, index=['Actual Negative', 'Actual Positive'], columns=['Predicted Negative', 'Predicted Positive'])
#heatmap
plt.figure(figsize=(8,6))
sns.heatmap(rf_df, annot=True, fmt='d', cmap='Blues')
plt.title('Random Forest Confusion Matrix')
plt.show()
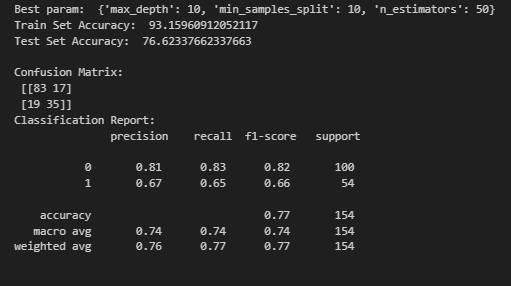
Observation:
The model’s training accuracy improved to 93.16%, while test accuracy increased to 76.62%, showing better generalization but still some overfitting.
The confusion matrix indicates that the model correctly classifies 83 non-diabetic and 35 diabetic patients, with fewer misclassifications compared to the previous model. However, 17 non-diabetic and 19 diabetic patients are still misclassified.
The classification report shows an improvement in detecting diabetic cases (precision = 0.67, recall = 0.65, f1-score = 0.66), meaning the model is now slightly better at identifying diabetes, though it still favors non-diabetic predictions (precision = 0.81, recall = 0.83).
Save The Model
The Random Forest classifier is the better-performing model; it will be saved using the pickle library and is useful in building our app. The standard scaler would also be saved to be used in the app When our user inputs details, the model would first scale the inputs before passing them into the model for prediction.
#import required library
import pickle
pickle.dump(rfc, open('model.pkl', 'wb'))
pickle.dump(scaler, open('scaler.pkl', 'wb'))
BUILD AND DEPLOY THE APP
Now, we would build and deploy the app using STREAMLIT.
import streamlit as st
import pickle
import numpy as np
import time
# Load the trained model and scaler
model = pickle.load(open('model.pkl', 'rb'))
scaler = pickle.load(open('scaler.pkl', 'rb'))
# Streamlit app styling
st.markdown(
"""
""",
unsafe_allow_html=True
)
# Title
st.markdown("""




































































































































































![[The AI Show Episode 142]: ChatGPT’s New Image Generator, Studio Ghibli Craze and Backlash, Gemini 2.5, OpenAI Academy, 4o Updates, Vibe Marketing & xAI Acquires X](https://www.marketingaiinstitute.com/hubfs/ep%20142%20cover.png)


































































































![[DEALS] The Premium Learn to Code Certification Bundle (97% off) & Other Deals Up To 98% Off – Offers End Soon!](https://www.javacodegeeks.com/wp-content/uploads/2012/12/jcg-logo.jpg)


![From drop-out to software architect with Jason Lengstorf [Podcast #167]](https://cdn.hashnode.com/res/hashnode/image/upload/v1743796461357/f3d19cd7-e6f5-4d7c-8bfc-eb974bc8da68.png?#)






























































































.png?#)





























_Christophe_Coat_Alamy.jpg?#)
 (1).webp?#)






























































































![iPhone 17 Pro Won't Feature Two-Toned Back [Gurman]](https://www.iclarified.com/images/news/96944/96944/96944-640.jpg)
![Tariffs Threaten Apple's $999 iPhone Price Point in the U.S. [Gurman]](https://www.iclarified.com/images/news/96943/96943/96943-640.jpg)