Dashboard for Researchers & Geneticists: Functional Requirements [System Design]
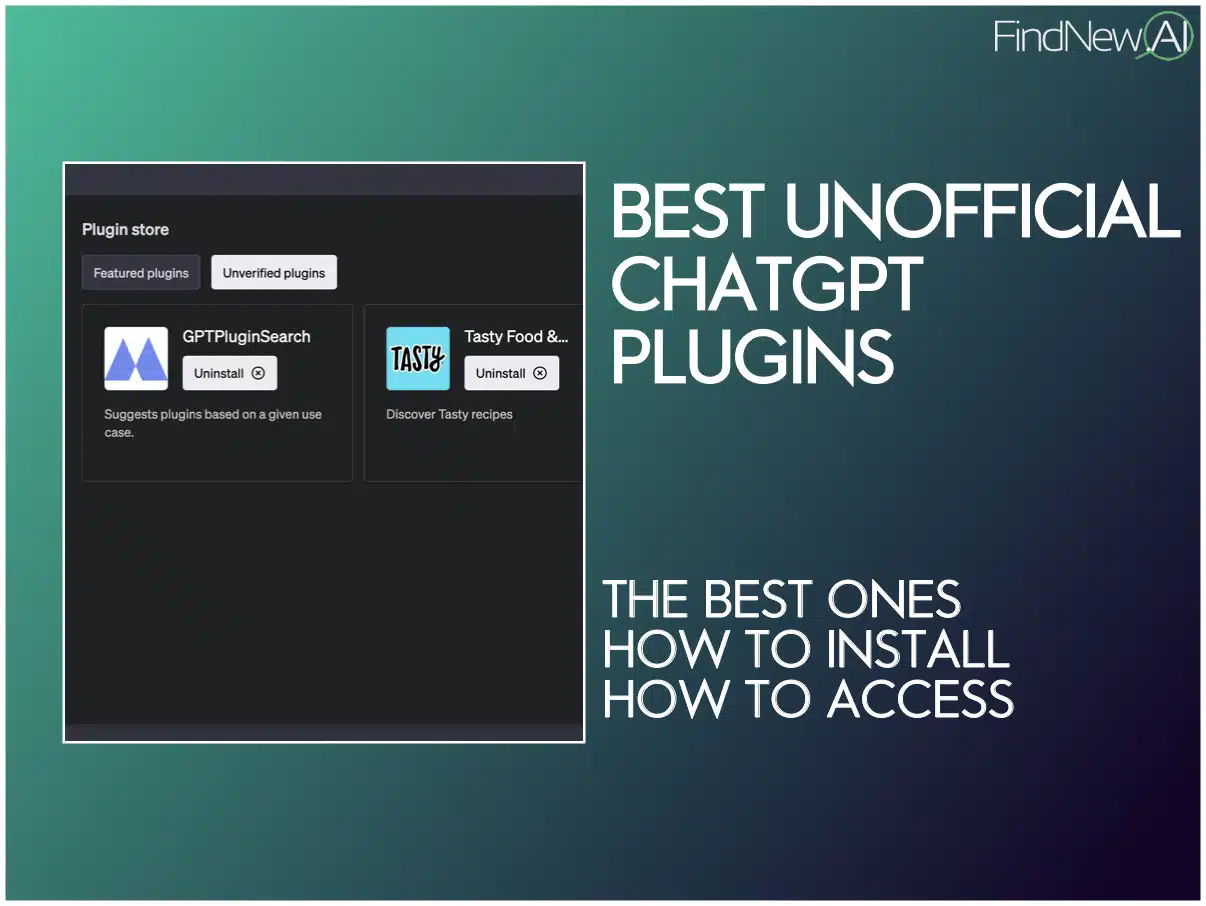
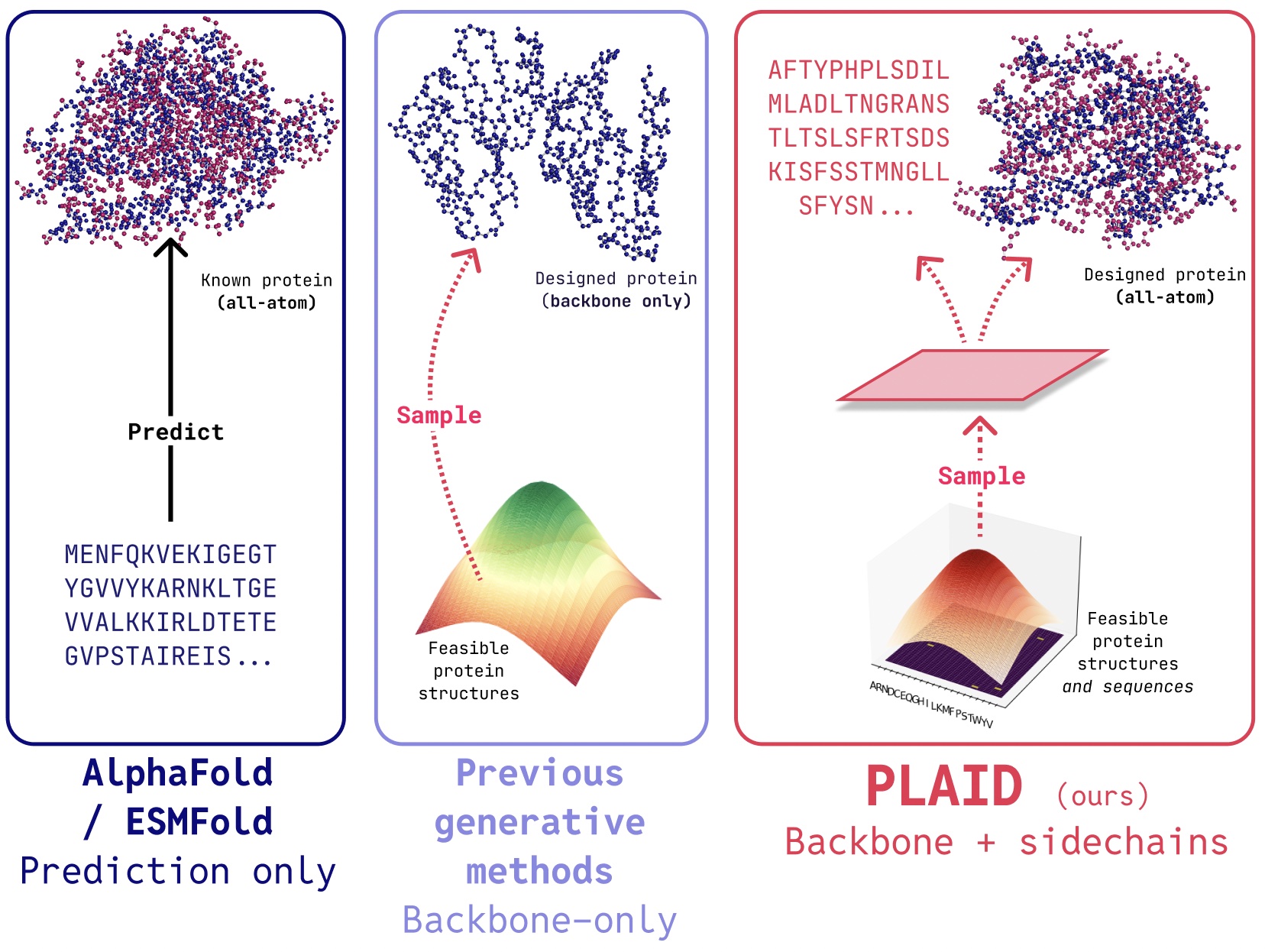
Overview of Features The system is designed for researchers and geneticists to analyze genetic/genomic data, with multi-level roles using Role-Based Access Control (RBAC). The system will have the following components: Genetic Data API: Built using Fast API, PostgreSQL for genetic data storage, and Redis for caching. User Management: Using NoSQL database for user data storage, with Redis for caching. ETL (Extract, Transform, Load): Using Apache Airflow to migrate legacy data to a modern SQL database with normalization, better relationships using ERD, indexing, etc. Authentication: Single Sign-On (SSO), OAuth, and Decentralized Identifier (DID) using Keri or similar (e.g., AT Protocol). We will also integrate web3 wallets such as MetaMask. Emilia AI: Integration with PubMed API, Nature Journal API, etc., for answering questions related to genetic data. RStudio Server and JupyterHub: Integration for researchers to create and test RShiny apps and Jupyter notebooks, with S3 for blob storage under user RBAC. Functional Requirements Genetic Data API Gateway API Endpoints: Create, Read, Update, Delete (CRUD) operations for genetic data. API documentation with OpenAPI/Swagger API for accessing (read access) genetic data from RStudio environment with limitations. Database: PostgreSQL for genetic data storage, with sharding & replica sets for scalability & high availability. Caching: Redis for caching genetic data. User Management Database: NoSQL database for user data storage. Caching: Redis for caching user data. RBAC with multi-level permissions: Researchers (focus): Full data analysis capabilities, access to computational environments Farmers: Limited access to relevant genetic data for their livestock, data entry capabilities, profile management Future Enhancement: Farmers will have a dedicated interface to input livestock data and manage their profiles, with tailored access to the Emilia AI assistant. Their AI interactions will be focused on practical applications such as optimal mating recommendations and basic genetic analysis for their specific herds, without access to the broader research capabilities available to geneticists and researchers. ETL Legacy Data Migration: Utilizing Apache Airflow to extract data from legacy systems and load it into a modern PostgreSQL database, enabling efficient data management and analysis. Data Normalization: Normalizing genetic data to optimize relationships and data integrity using Entity-Relationship Diagrams (ERD), ensuring a robust and scalable database design. Indexing: Implementing indexing strategies to facilitate efficient data retrieval and querying, enhancing overall system performance. Authentication Single Sign-On (SSO): Implementing SSO for streamlined user authentication and access control. OAuth: Utilizing OAuth for secure authorization and authentication. Decentralized Identifier (DID): Integrating AT Protocol for decentralized identity management, enabling users to control their digital identity. Web3 & MetaMask Integration: Incorporating Web3 technologies and MetaMask for seamless blockchain-based authentication and interaction, ensuring a secure and decentralized user experience. Emilia AI Assistant Genetic/Genomic Database Integration: Integrating genetic and genomic databases to provide accurate and informed answers. LLM & RAG: Utilizing Large Language Models (LLM), Natural Language Processing (NLP - e.g., Lanchain) and Retrieval-Augmented Generation (RAG - e.g., Amazon Bedrock) to optimize answers based on genetic/genomic data and research publications. Journal APIs: Leveraging APIs from reputable journals (e.g., PubMed API), Nature API) to access the latest research publications and stay up-to-date with the latest findings. Downloaded Articles/Publications: In cases where APIs are not available, utilizing downloaded articles and publications as a starting point to ensure comprehensive coverage and insights. DAG for Heritability: Implementing Directed Acyclic Graphs (DAG) to estimate heritability and identify genetic relationships. Users can ask Emilia AI questions such as: What are the known genetic factors contributing to a certain trait? What are the genetic factors influencing milk yield in dairy cattle? What is the estimated breeding value (EBV) for a specific trait in a particular breed of cattle? Can you suggest a suitable mate for cow #XXX based on its genetic profile and breeding goals? Future Enhancement: Custom Models: Developing custom models for advanced analyses, such as: Genome-Wide Association Studies (GWAS) Gene expression analysis Re-engineering Analysis Tools: Integrating and re-engineering popular analysis tools, such as: PLINK: Whole genome association analysis toolset User Query → NLP Parser → Intent Classification → PLINK Command Generator → PLINK Execution → Result Parser → Context Enhanc
![Dashboard for Researchers & Geneticists: Functional Requirements [System Design]](https://media2.dev.to/dynamic/image/width=800%2Cheight=%2Cfit=scale-down%2Cgravity=auto%2Cformat=auto/https%3A%2F%2Fdev-to-uploads.s3.amazonaws.com%2Fuploads%2Farticles%2F37i2spw6bi3fa17c5xqe.png)
Overview of Features
The system is designed for researchers and geneticists to analyze genetic/genomic data, with multi-level roles using Role-Based Access Control (RBAC). The system will have the following components:
Genetic Data API: Built using Fast API, PostgreSQL for genetic data storage, and Redis for caching.
User Management: Using NoSQL database for user data storage, with Redis for caching.
ETL (Extract, Transform, Load): Using Apache Airflow to migrate legacy data to a modern SQL database with normalization, better relationships using ERD, indexing, etc.
Authentication: Single Sign-On (SSO), OAuth, and Decentralized Identifier (DID) using Keri or similar (e.g., AT Protocol). We will also integrate web3 wallets such as MetaMask.
Emilia AI: Integration with PubMed API, Nature Journal API, etc., for answering questions related to genetic data.
RStudio Server and JupyterHub: Integration for researchers to create and test RShiny apps and Jupyter notebooks, with S3 for blob storage under user RBAC.
Functional Requirements
Genetic Data API Gateway
-
API Endpoints:
- Create, Read, Update, Delete (CRUD) operations for genetic data.
- API documentation with OpenAPI/Swagger
- API for accessing (read access) genetic data from RStudio environment with limitations.
Database: PostgreSQL for genetic data storage, with sharding & replica sets for scalability & high availability.
Caching: Redis for caching genetic data.
User Management
Database: NoSQL database for user data storage.
Caching: Redis for caching user data.
-
RBAC with multi-level permissions:
- Researchers (focus): Full data analysis capabilities, access to computational environments
- Farmers: Limited access to relevant genetic data for their livestock, data entry capabilities, profile management
Future Enhancement: Farmers will have a dedicated interface to input livestock data and manage their profiles, with tailored access to the Emilia AI assistant. Their AI interactions will be focused on practical applications such as optimal mating recommendations and basic genetic analysis for their specific herds, without access to the broader research capabilities available to geneticists and researchers.
ETL
Legacy Data Migration: Utilizing Apache Airflow to extract data from legacy systems and load it into a modern PostgreSQL database, enabling efficient data management and analysis.
Data Normalization: Normalizing genetic data to optimize relationships and data integrity using Entity-Relationship Diagrams (ERD), ensuring a robust and scalable database design.
Indexing: Implementing indexing strategies to facilitate efficient data retrieval and querying, enhancing overall system performance.
Authentication
Single Sign-On (SSO): Implementing SSO for streamlined user authentication and access control.
OAuth: Utilizing OAuth for secure authorization and authentication.
Decentralized Identifier (DID): Integrating AT Protocol for decentralized identity management, enabling users to control their digital identity.
Web3 & MetaMask Integration: Incorporating Web3 technologies and MetaMask for seamless blockchain-based authentication and interaction, ensuring a secure and decentralized user experience.
Emilia AI Assistant
Genetic/Genomic Database Integration: Integrating genetic and genomic databases to provide accurate and informed answers.
LLM & RAG: Utilizing Large Language Models (LLM), Natural Language Processing (NLP - e.g., Lanchain) and Retrieval-Augmented Generation (RAG - e.g., Amazon Bedrock) to optimize answers based on genetic/genomic data and research publications.
Journal APIs: Leveraging APIs from reputable journals (e.g., PubMed API), Nature API) to access the latest research publications and stay up-to-date with the latest findings.
Downloaded Articles/Publications: In cases where APIs are not available, utilizing downloaded articles and publications as a starting point to ensure comprehensive coverage and insights.
DAG for Heritability: Implementing Directed Acyclic Graphs (DAG) to estimate heritability and identify genetic relationships.
-
Users can ask Emilia AI questions such as:
- What are the known genetic factors contributing to a certain trait?
- What are the genetic factors influencing milk yield in dairy cattle?
- What is the estimated breeding value (EBV) for a specific trait in a particular breed of cattle?
- Can you suggest a suitable mate for cow #XXX based on its genetic profile and breeding goals?
-
Future Enhancement:
- Custom Models: Developing custom models for advanced analyses, such as:
- Genome-Wide Association Studies (GWAS)
- Gene expression analysis
- Re-engineering Analysis Tools: Integrating and re-engineering popular analysis tools, such as:
- Custom Models: Developing custom models for advanced analyses, such as:
RStudio Server and JupyterHub


































































































































































![[The AI Show Episode 144]: ChatGPT’s New Memory, Shopify CEO’s Leaked “AI First” Memo, Google Cloud Next Releases, o3 and o4-mini Coming Soon & Llama 4’s Rocky Launch](https://www.marketingaiinstitute.com/hubfs/ep%20144%20cover.png)





































































































![[DEALS] The All-in-One Microsoft Office Pro 2019 for Windows: Lifetime License + Windows 11 Pro Bundle (89% off) & Other Deals Up To 98% Off](https://www.javacodegeeks.com/wp-content/uploads/2012/12/jcg-logo.jpg)
























![Is this too much for a modular monolith system? [closed]](https://i.sstatic.net/pYL1nsfg.png)
















































































































_Andreas_Prott_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)
























































































![What features do you get with Gemini Advanced? [April 2025]](https://i0.wp.com/9to5google.com/wp-content/uploads/sites/4/2024/02/gemini-advanced-cover.jpg?resize=1200%2C628&quality=82&strip=all&ssl=1)













![Apple Shares Official Trailer for 'Long Way Home' Starring Ewan McGregor and Charley Boorman [Video]](https://www.iclarified.com/images/news/97069/97069/97069-640.jpg)
![Apple Watch Series 10 Back On Sale for $299! [Lowest Price Ever]](https://www.iclarified.com/images/news/96657/96657/96657-640.jpg)
![EU Postpones Apple App Store Fines Amid Tariff Negotiations [Report]](https://www.iclarified.com/images/news/97068/97068/97068-640.jpg)
![Apple Slips to Fifth in China's Smartphone Market with 9% Decline [Report]](https://www.iclarified.com/images/news/97065/97065/97065-640.jpg)